
Oryctes borbonicus
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Panarthropoda; Arthropoda; Mandibulata; Pancrustacea; Hexapoda; Insecta; Dicondylia; Pterygota; Neoptera; Endopterygota; Coleoptera; Polyphaga; Scarabaeiformia; Scarabaeoidea; Scarabaeidae; Dynastinae;
Average proteome isoelectric point is 6.68
Get precalculated fractions of proteins
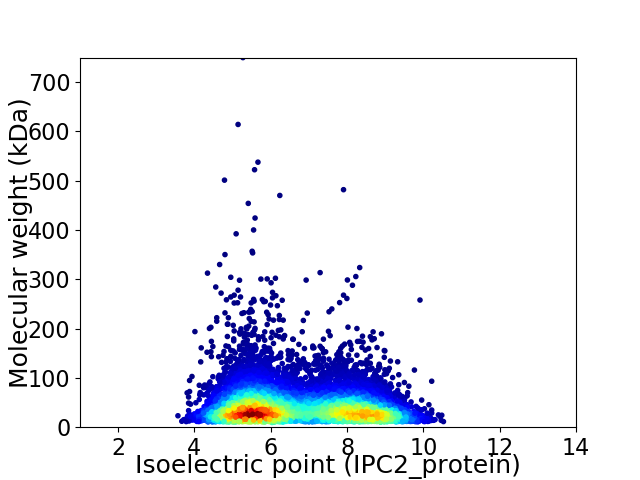
Virtual 2D-PAGE plot for 8820 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
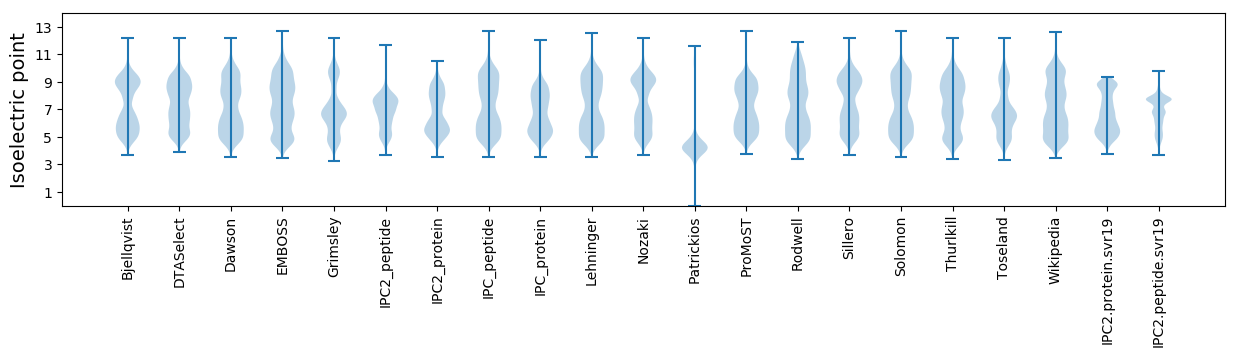
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0T6BA91|A0A0T6BA91_9SCAR Uncharacterized protein (Fragment) OS=Oryctes borbonicus OX=1629725 GN=AMK59_1340 PE=4 SV=1
VV1 pKa = 5.79EE2 pKa = 4.18TADD5 pKa = 3.7SLRR8 pKa = 11.84ATWEE12 pKa = 3.85APLNSEE18 pKa = 4.31EE19 pKa = 4.78CGVAYY24 pKa = 9.52EE25 pKa = 4.24VIYY28 pKa = 10.69EE29 pKa = 3.92NEE31 pKa = 4.06ILGSSAVEE39 pKa = 4.25VVGLAHH45 pKa = 7.27DD46 pKa = 4.39FNIEE50 pKa = 4.77LIPCTDD56 pKa = 3.31YY57 pKa = 11.24TITVTPISFTGSKK70 pKa = 8.31GTPVSQEE77 pKa = 3.63VTPSIRR83 pKa = 11.84VGSVNDD89 pKa = 4.93LIAADD94 pKa = 3.82VGDD97 pKa = 4.24SVVLSWSTATVGVCPLVYY115 pKa = 9.9IVNYY119 pKa = 9.96AVNDD123 pKa = 3.7VQGEE127 pKa = 4.14EE128 pKa = 4.52LEE130 pKa = 4.4TTEE133 pKa = 5.14VSVTFNRR140 pKa = 11.84IPCSDD145 pKa = 3.37SQFSVKK151 pKa = 10.13VLSEE155 pKa = 4.55GIVGPVAVANLEE167 pKa = 4.29LGSADD172 pKa = 3.35INSITQVEE180 pKa = 4.55GLNLDD185 pKa = 3.61GTILSWSPPVTVGLCEE201 pKa = 3.3IVYY204 pKa = 10.32IVDD207 pKa = 3.34IEE209 pKa = 4.46NTFGLQQTATRR220 pKa = 11.84DD221 pKa = 3.46LSIDD225 pKa = 3.99LEE227 pKa = 4.86LIPCSRR233 pKa = 11.84NLFVVRR239 pKa = 11.84GSANGITGEE248 pKa = 3.89PATIEE253 pKa = 4.15EE254 pKa = 4.45YY255 pKa = 11.27APIAEE260 pKa = 4.83LAPPTHH266 pKa = 6.35VRR268 pKa = 11.84TKK270 pKa = 10.32PAFTSADD277 pKa = 2.93ITLQGQAISEE287 pKa = 4.31NICRR291 pKa = 11.84VEE293 pKa = 3.96TAILEE298 pKa = 4.39CSDD301 pKa = 2.89ISGGVIEE308 pKa = 5.52RR309 pKa = 11.84EE310 pKa = 3.79IEE312 pKa = 4.26VMDD315 pKa = 3.4NTPRR319 pKa = 11.84IFTVLLDD326 pKa = 4.84DD327 pKa = 5.54LEE329 pKa = 4.67QDD331 pKa = 3.81TIYY334 pKa = 10.6TCVARR339 pKa = 11.84LEE341 pKa = 4.45DD342 pKa = 3.58SAAGNEE348 pKa = 4.25FSFRR352 pKa = 11.84TVPLVEE358 pKa = 4.65KK359 pKa = 10.61DD360 pKa = 4.53LIISDD365 pKa = 3.33ITEE368 pKa = 4.11GGFLVSWIEE377 pKa = 4.39DD378 pKa = 3.33YY379 pKa = 11.16GQQFVIGYY387 pKa = 7.75EE388 pKa = 3.91LTIEE392 pKa = 4.35SLGPAHH398 pKa = 7.37RR399 pKa = 11.84VPDD402 pKa = 4.12GCSVDD407 pKa = 3.69TTVTTVSLSNEE418 pKa = 4.07EE419 pKa = 4.06LSSAFVGGLPDD430 pKa = 3.18YY431 pKa = 10.5AYY433 pKa = 10.21RR434 pKa = 11.84VVLTTYY440 pKa = 8.4YY441 pKa = 10.66TLGLSLQSNTASARR455 pKa = 11.84TLAGVPSLPEE465 pKa = 3.7NLVLDD470 pKa = 4.68FVSGSTEE477 pKa = 3.91AYY479 pKa = 9.01NVSAILSWSSPCEE492 pKa = 3.78LNGGLRR498 pKa = 11.84GFGFNITARR507 pKa = 11.84FSLGGVEE514 pKa = 4.84QSVTVNLGVDD524 pKa = 3.64EE525 pKa = 5.64DD526 pKa = 4.6DD527 pKa = 3.93VNAVISKK534 pKa = 8.8TITDD538 pKa = 3.55INPFFRR544 pKa = 11.84YY545 pKa = 8.61EE546 pKa = 3.79VSVNVIAADD555 pKa = 4.05DD556 pKa = 4.48GPNAVLGFIPPAACVCDD573 pKa = 3.59AAA575 pKa = 5.95
VV1 pKa = 5.79EE2 pKa = 4.18TADD5 pKa = 3.7SLRR8 pKa = 11.84ATWEE12 pKa = 3.85APLNSEE18 pKa = 4.31EE19 pKa = 4.78CGVAYY24 pKa = 9.52EE25 pKa = 4.24VIYY28 pKa = 10.69EE29 pKa = 3.92NEE31 pKa = 4.06ILGSSAVEE39 pKa = 4.25VVGLAHH45 pKa = 7.27DD46 pKa = 4.39FNIEE50 pKa = 4.77LIPCTDD56 pKa = 3.31YY57 pKa = 11.24TITVTPISFTGSKK70 pKa = 8.31GTPVSQEE77 pKa = 3.63VTPSIRR83 pKa = 11.84VGSVNDD89 pKa = 4.93LIAADD94 pKa = 3.82VGDD97 pKa = 4.24SVVLSWSTATVGVCPLVYY115 pKa = 9.9IVNYY119 pKa = 9.96AVNDD123 pKa = 3.7VQGEE127 pKa = 4.14EE128 pKa = 4.52LEE130 pKa = 4.4TTEE133 pKa = 5.14VSVTFNRR140 pKa = 11.84IPCSDD145 pKa = 3.37SQFSVKK151 pKa = 10.13VLSEE155 pKa = 4.55GIVGPVAVANLEE167 pKa = 4.29LGSADD172 pKa = 3.35INSITQVEE180 pKa = 4.55GLNLDD185 pKa = 3.61GTILSWSPPVTVGLCEE201 pKa = 3.3IVYY204 pKa = 10.32IVDD207 pKa = 3.34IEE209 pKa = 4.46NTFGLQQTATRR220 pKa = 11.84DD221 pKa = 3.46LSIDD225 pKa = 3.99LEE227 pKa = 4.86LIPCSRR233 pKa = 11.84NLFVVRR239 pKa = 11.84GSANGITGEE248 pKa = 3.89PATIEE253 pKa = 4.15EE254 pKa = 4.45YY255 pKa = 11.27APIAEE260 pKa = 4.83LAPPTHH266 pKa = 6.35VRR268 pKa = 11.84TKK270 pKa = 10.32PAFTSADD277 pKa = 2.93ITLQGQAISEE287 pKa = 4.31NICRR291 pKa = 11.84VEE293 pKa = 3.96TAILEE298 pKa = 4.39CSDD301 pKa = 2.89ISGGVIEE308 pKa = 5.52RR309 pKa = 11.84EE310 pKa = 3.79IEE312 pKa = 4.26VMDD315 pKa = 3.4NTPRR319 pKa = 11.84IFTVLLDD326 pKa = 4.84DD327 pKa = 5.54LEE329 pKa = 4.67QDD331 pKa = 3.81TIYY334 pKa = 10.6TCVARR339 pKa = 11.84LEE341 pKa = 4.45DD342 pKa = 3.58SAAGNEE348 pKa = 4.25FSFRR352 pKa = 11.84TVPLVEE358 pKa = 4.65KK359 pKa = 10.61DD360 pKa = 4.53LIISDD365 pKa = 3.33ITEE368 pKa = 4.11GGFLVSWIEE377 pKa = 4.39DD378 pKa = 3.33YY379 pKa = 11.16GQQFVIGYY387 pKa = 7.75EE388 pKa = 3.91LTIEE392 pKa = 4.35SLGPAHH398 pKa = 7.37RR399 pKa = 11.84VPDD402 pKa = 4.12GCSVDD407 pKa = 3.69TTVTTVSLSNEE418 pKa = 4.07EE419 pKa = 4.06LSSAFVGGLPDD430 pKa = 3.18YY431 pKa = 10.5AYY433 pKa = 10.21RR434 pKa = 11.84VVLTTYY440 pKa = 8.4YY441 pKa = 10.66TLGLSLQSNTASARR455 pKa = 11.84TLAGVPSLPEE465 pKa = 3.7NLVLDD470 pKa = 4.68FVSGSTEE477 pKa = 3.91AYY479 pKa = 9.01NVSAILSWSSPCEE492 pKa = 3.78LNGGLRR498 pKa = 11.84GFGFNITARR507 pKa = 11.84FSLGGVEE514 pKa = 4.84QSVTVNLGVDD524 pKa = 3.64EE525 pKa = 5.64DD526 pKa = 4.6DD527 pKa = 3.93VNAVISKK534 pKa = 8.8TITDD538 pKa = 3.55INPFFRR544 pKa = 11.84YY545 pKa = 8.61EE546 pKa = 3.79VSVNVIAADD555 pKa = 4.05DD556 pKa = 4.48GPNAVLGFIPPAACVCDD573 pKa = 3.59AAA575 pKa = 5.95
Molecular weight: 61.23 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0T6B3U6|A0A0T6B3U6_9SCAR LIM domain containing protein OS=Oryctes borbonicus OX=1629725 GN=AMK59_6067 PE=4 SV=1
RR1 pKa = 7.08NGGRR5 pKa = 11.84AKK7 pKa = 10.35HH8 pKa = 5.76GRR10 pKa = 11.84GHH12 pKa = 5.99VKK14 pKa = 10.0PVRR17 pKa = 11.84CTNCARR23 pKa = 11.84CVPKK27 pKa = 10.77DD28 pKa = 3.35KK29 pKa = 10.79AIKK32 pKa = 10.32KK33 pKa = 8.63FVIRR37 pKa = 11.84NIVEE41 pKa = 3.73AAAVRR46 pKa = 11.84DD47 pKa = 3.62ITDD50 pKa = 3.15ASVYY54 pKa = 9.41TSYY57 pKa = 11.64VLPKK61 pKa = 10.2LYY63 pKa = 10.77AKK65 pKa = 10.42LHH67 pKa = 5.53YY68 pKa = 10.0CVSCAIHH75 pKa = 6.27SKK77 pKa = 8.47VVRR80 pKa = 11.84NRR82 pKa = 11.84SKK84 pKa = 11.01EE85 pKa = 3.92EE86 pKa = 3.78RR87 pKa = 11.84KK88 pKa = 10.18KK89 pKa = 9.95RR90 pKa = 11.84TPPARR95 pKa = 11.84NFPGRR100 pKa = 11.84DD101 pKa = 3.11NVRR104 pKa = 11.84VVQQ107 pKa = 3.84
RR1 pKa = 7.08NGGRR5 pKa = 11.84AKK7 pKa = 10.35HH8 pKa = 5.76GRR10 pKa = 11.84GHH12 pKa = 5.99VKK14 pKa = 10.0PVRR17 pKa = 11.84CTNCARR23 pKa = 11.84CVPKK27 pKa = 10.77DD28 pKa = 3.35KK29 pKa = 10.79AIKK32 pKa = 10.32KK33 pKa = 8.63FVIRR37 pKa = 11.84NIVEE41 pKa = 3.73AAAVRR46 pKa = 11.84DD47 pKa = 3.62ITDD50 pKa = 3.15ASVYY54 pKa = 9.41TSYY57 pKa = 11.64VLPKK61 pKa = 10.2LYY63 pKa = 10.77AKK65 pKa = 10.42LHH67 pKa = 5.53YY68 pKa = 10.0CVSCAIHH75 pKa = 6.27SKK77 pKa = 8.47VVRR80 pKa = 11.84NRR82 pKa = 11.84SKK84 pKa = 11.01EE85 pKa = 3.92EE86 pKa = 3.78RR87 pKa = 11.84KK88 pKa = 10.18KK89 pKa = 9.95RR90 pKa = 11.84TPPARR95 pKa = 11.84NFPGRR100 pKa = 11.84DD101 pKa = 3.11NVRR104 pKa = 11.84VVQQ107 pKa = 3.84
Molecular weight: 12.13 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3587004 |
99 |
6462 |
406.7 |
46.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.598 ± 0.025 | 2.059 ± 0.029 |
5.564 ± 0.02 | 6.818 ± 0.037 |
3.856 ± 0.022 | 5.195 ± 0.037 |
2.424 ± 0.012 | 6.284 ± 0.021 |
7.037 ± 0.036 | 9.127 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.237 ± 0.013 | 5.566 ± 0.024 |
4.946 ± 0.035 | 4.227 ± 0.025 |
4.977 ± 0.024 | 7.821 ± 0.034 |
5.882 ± 0.021 | 5.996 ± 0.022 |
1.018 ± 0.009 | 3.368 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
