
Capybara microvirus Cap1_SP_81
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
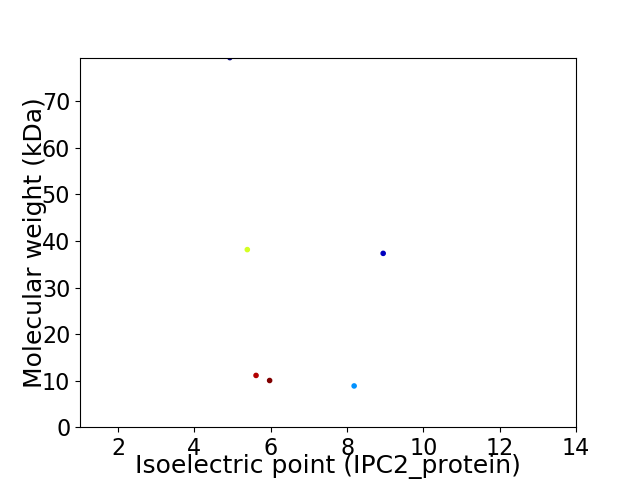
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W496|A0A4P8W496_9VIRU Minor capsid protein OS=Capybara microvirus Cap1_SP_81 OX=2584792 PE=4 SV=1
MM1 pKa = 7.38SLSKK5 pKa = 9.94TIGGDD10 pKa = 3.04RR11 pKa = 11.84LGAGNKK17 pKa = 9.1QKK19 pKa = 10.51VHH21 pKa = 4.94LHH23 pKa = 5.14NYY25 pKa = 7.97EE26 pKa = 3.87RR27 pKa = 11.84ATFNLDD33 pKa = 2.88KK34 pKa = 10.56VVRR37 pKa = 11.84MSMAPGILYY46 pKa = 9.07PFYY49 pKa = 11.21QNVALLGDD57 pKa = 3.5TWDD60 pKa = 3.5ININSLVRR68 pKa = 11.84TIPTIGPLFGTFKK81 pKa = 9.94MQIDD85 pKa = 3.98FFFYY89 pKa = 10.31PIRR92 pKa = 11.84LAQGLLHH99 pKa = 6.65NNPLNLGMEE108 pKa = 4.22TDD110 pKa = 4.34KK111 pKa = 11.27IKK113 pKa = 10.92LPQIEE118 pKa = 4.97IEE120 pKa = 4.03TDD122 pKa = 3.36TQKK125 pKa = 10.81SEE127 pKa = 4.51RR128 pKa = 11.84NGNNWQINQSSLAAYY143 pKa = 10.09LGIRR147 pKa = 11.84GIGAPTWAEE156 pKa = 3.55QSGEE160 pKa = 4.25IIWVNPTIKK169 pKa = 10.31RR170 pKa = 11.84KK171 pKa = 9.85FNATTFLAYY180 pKa = 10.32ADD182 pKa = 3.88IFKK185 pKa = 10.61SYY187 pKa = 10.34YY188 pKa = 10.53ANKK191 pKa = 10.06QEE193 pKa = 4.27DD194 pKa = 3.52NAYY197 pKa = 10.88VIGQEE202 pKa = 4.14LGKK205 pKa = 9.6EE206 pKa = 4.09HH207 pKa = 7.16QEE209 pKa = 3.99IPDD212 pKa = 3.64WLINNYY218 pKa = 9.71LFSFGNNNGQYY229 pKa = 10.65CYY231 pKa = 11.18LNWNDD236 pKa = 5.25NEE238 pKa = 4.43KK239 pKa = 10.02QWEE242 pKa = 4.52CNLQFAPTYY251 pKa = 10.04IKK253 pKa = 10.74YY254 pKa = 10.28EE255 pKa = 4.12RR256 pKa = 11.84FDD258 pKa = 3.86KK259 pKa = 11.01DD260 pKa = 3.11HH261 pKa = 6.9SALEE265 pKa = 4.11VNMWLMSRR273 pKa = 11.84ISITTKK279 pKa = 9.95GKK281 pKa = 8.43TYY283 pKa = 10.85KK284 pKa = 10.62LPEE287 pKa = 5.07LINLEE292 pKa = 4.3VISDD296 pKa = 4.74KK297 pKa = 10.52ITITTPSEE305 pKa = 3.83YY306 pKa = 10.19EE307 pKa = 3.75FYY309 pKa = 11.28GNYY312 pKa = 8.42VQYY315 pKa = 10.8HH316 pKa = 7.1IINNSIFGGDD326 pKa = 3.74TIVEE330 pKa = 4.31TNTYY334 pKa = 11.0NEE336 pKa = 4.05TSKK339 pKa = 11.17LKK341 pKa = 11.08LIDD344 pKa = 4.22FPLKK348 pKa = 10.9NIDD351 pKa = 3.42DD352 pKa = 3.76ARR354 pKa = 11.84YY355 pKa = 9.71IILGHH360 pKa = 5.5NQPNEE365 pKa = 3.83ALSINEE371 pKa = 4.64DD372 pKa = 4.22LNFLPYY378 pKa = 10.31SQWSGKK384 pKa = 7.22DD385 pKa = 3.05TKK387 pKa = 11.21NRR389 pKa = 11.84TNNYY393 pKa = 7.31WPHH396 pKa = 6.22NGLFIKK402 pKa = 9.62TYY404 pKa = 10.51QSDD407 pKa = 4.18LFNNWVKK414 pKa = 10.37TDD416 pKa = 5.22FITGDD421 pKa = 3.24NGIQEE426 pKa = 4.06ITKK429 pKa = 9.83IDD431 pKa = 3.72TTDD434 pKa = 3.38GLILDD439 pKa = 5.02EE440 pKa = 5.15LNLMQKK446 pKa = 9.59VYY448 pKa = 11.57NMLNRR453 pKa = 11.84VAISGGTYY461 pKa = 9.73EE462 pKa = 5.9DD463 pKa = 4.18YY464 pKa = 11.64LEE466 pKa = 4.17VQYY469 pKa = 11.15GVSALRR475 pKa = 11.84LEE477 pKa = 4.27EE478 pKa = 4.07TPVYY482 pKa = 10.02IGGYY486 pKa = 6.15STEE489 pKa = 3.93IDD491 pKa = 3.71FEE493 pKa = 4.58EE494 pKa = 4.64VTSNAASNATGEE506 pKa = 4.15IQNLGSLAGKK516 pKa = 9.85GINRR520 pKa = 11.84SPKK523 pKa = 9.91GGRR526 pKa = 11.84ISYY529 pKa = 9.46KK530 pKa = 9.14PKK532 pKa = 10.17EE533 pKa = 3.9IGFIMGICSLTPRR546 pKa = 11.84IDD548 pKa = 3.41YY549 pKa = 10.75SQGNKK554 pKa = 7.84FWEE557 pKa = 4.47TEE559 pKa = 3.79IEE561 pKa = 4.21TLADD565 pKa = 3.3FHH567 pKa = 7.77VPAMDD572 pKa = 5.16GIGYY576 pKa = 9.56QDD578 pKa = 5.96LITEE582 pKa = 3.94QMAYY586 pKa = 10.36FDD588 pKa = 4.42TLILPDD594 pKa = 3.8GEE596 pKa = 4.63IIKK599 pKa = 10.47HH600 pKa = 5.01SAGKK604 pKa = 9.87IPAWLNYY611 pKa = 6.66MTDD614 pKa = 3.13VNEE617 pKa = 4.49TYY619 pKa = 11.45GNFADD624 pKa = 3.94NDD626 pKa = 3.9EE627 pKa = 4.81TSGQMWMTLNRR638 pKa = 11.84NYY640 pKa = 10.56EE641 pKa = 4.03RR642 pKa = 11.84TNDD645 pKa = 4.33GIGDD649 pKa = 4.18LTTYY653 pKa = 10.58IDD655 pKa = 3.63PTKK658 pKa = 10.63YY659 pKa = 10.59NYY661 pKa = 10.92AFAVKK666 pKa = 10.23DD667 pKa = 3.72LEE669 pKa = 4.43AQNFWAQIGMNIVARR684 pKa = 11.84RR685 pKa = 11.84KK686 pKa = 9.14MSAKK690 pKa = 10.1QIPNLL695 pKa = 3.74
MM1 pKa = 7.38SLSKK5 pKa = 9.94TIGGDD10 pKa = 3.04RR11 pKa = 11.84LGAGNKK17 pKa = 9.1QKK19 pKa = 10.51VHH21 pKa = 4.94LHH23 pKa = 5.14NYY25 pKa = 7.97EE26 pKa = 3.87RR27 pKa = 11.84ATFNLDD33 pKa = 2.88KK34 pKa = 10.56VVRR37 pKa = 11.84MSMAPGILYY46 pKa = 9.07PFYY49 pKa = 11.21QNVALLGDD57 pKa = 3.5TWDD60 pKa = 3.5ININSLVRR68 pKa = 11.84TIPTIGPLFGTFKK81 pKa = 9.94MQIDD85 pKa = 3.98FFFYY89 pKa = 10.31PIRR92 pKa = 11.84LAQGLLHH99 pKa = 6.65NNPLNLGMEE108 pKa = 4.22TDD110 pKa = 4.34KK111 pKa = 11.27IKK113 pKa = 10.92LPQIEE118 pKa = 4.97IEE120 pKa = 4.03TDD122 pKa = 3.36TQKK125 pKa = 10.81SEE127 pKa = 4.51RR128 pKa = 11.84NGNNWQINQSSLAAYY143 pKa = 10.09LGIRR147 pKa = 11.84GIGAPTWAEE156 pKa = 3.55QSGEE160 pKa = 4.25IIWVNPTIKK169 pKa = 10.31RR170 pKa = 11.84KK171 pKa = 9.85FNATTFLAYY180 pKa = 10.32ADD182 pKa = 3.88IFKK185 pKa = 10.61SYY187 pKa = 10.34YY188 pKa = 10.53ANKK191 pKa = 10.06QEE193 pKa = 4.27DD194 pKa = 3.52NAYY197 pKa = 10.88VIGQEE202 pKa = 4.14LGKK205 pKa = 9.6EE206 pKa = 4.09HH207 pKa = 7.16QEE209 pKa = 3.99IPDD212 pKa = 3.64WLINNYY218 pKa = 9.71LFSFGNNNGQYY229 pKa = 10.65CYY231 pKa = 11.18LNWNDD236 pKa = 5.25NEE238 pKa = 4.43KK239 pKa = 10.02QWEE242 pKa = 4.52CNLQFAPTYY251 pKa = 10.04IKK253 pKa = 10.74YY254 pKa = 10.28EE255 pKa = 4.12RR256 pKa = 11.84FDD258 pKa = 3.86KK259 pKa = 11.01DD260 pKa = 3.11HH261 pKa = 6.9SALEE265 pKa = 4.11VNMWLMSRR273 pKa = 11.84ISITTKK279 pKa = 9.95GKK281 pKa = 8.43TYY283 pKa = 10.85KK284 pKa = 10.62LPEE287 pKa = 5.07LINLEE292 pKa = 4.3VISDD296 pKa = 4.74KK297 pKa = 10.52ITITTPSEE305 pKa = 3.83YY306 pKa = 10.19EE307 pKa = 3.75FYY309 pKa = 11.28GNYY312 pKa = 8.42VQYY315 pKa = 10.8HH316 pKa = 7.1IINNSIFGGDD326 pKa = 3.74TIVEE330 pKa = 4.31TNTYY334 pKa = 11.0NEE336 pKa = 4.05TSKK339 pKa = 11.17LKK341 pKa = 11.08LIDD344 pKa = 4.22FPLKK348 pKa = 10.9NIDD351 pKa = 3.42DD352 pKa = 3.76ARR354 pKa = 11.84YY355 pKa = 9.71IILGHH360 pKa = 5.5NQPNEE365 pKa = 3.83ALSINEE371 pKa = 4.64DD372 pKa = 4.22LNFLPYY378 pKa = 10.31SQWSGKK384 pKa = 7.22DD385 pKa = 3.05TKK387 pKa = 11.21NRR389 pKa = 11.84TNNYY393 pKa = 7.31WPHH396 pKa = 6.22NGLFIKK402 pKa = 9.62TYY404 pKa = 10.51QSDD407 pKa = 4.18LFNNWVKK414 pKa = 10.37TDD416 pKa = 5.22FITGDD421 pKa = 3.24NGIQEE426 pKa = 4.06ITKK429 pKa = 9.83IDD431 pKa = 3.72TTDD434 pKa = 3.38GLILDD439 pKa = 5.02EE440 pKa = 5.15LNLMQKK446 pKa = 9.59VYY448 pKa = 11.57NMLNRR453 pKa = 11.84VAISGGTYY461 pKa = 9.73EE462 pKa = 5.9DD463 pKa = 4.18YY464 pKa = 11.64LEE466 pKa = 4.17VQYY469 pKa = 11.15GVSALRR475 pKa = 11.84LEE477 pKa = 4.27EE478 pKa = 4.07TPVYY482 pKa = 10.02IGGYY486 pKa = 6.15STEE489 pKa = 3.93IDD491 pKa = 3.71FEE493 pKa = 4.58EE494 pKa = 4.64VTSNAASNATGEE506 pKa = 4.15IQNLGSLAGKK516 pKa = 9.85GINRR520 pKa = 11.84SPKK523 pKa = 9.91GGRR526 pKa = 11.84ISYY529 pKa = 9.46KK530 pKa = 9.14PKK532 pKa = 10.17EE533 pKa = 3.9IGFIMGICSLTPRR546 pKa = 11.84IDD548 pKa = 3.41YY549 pKa = 10.75SQGNKK554 pKa = 7.84FWEE557 pKa = 4.47TEE559 pKa = 3.79IEE561 pKa = 4.21TLADD565 pKa = 3.3FHH567 pKa = 7.77VPAMDD572 pKa = 5.16GIGYY576 pKa = 9.56QDD578 pKa = 5.96LITEE582 pKa = 3.94QMAYY586 pKa = 10.36FDD588 pKa = 4.42TLILPDD594 pKa = 3.8GEE596 pKa = 4.63IIKK599 pKa = 10.47HH600 pKa = 5.01SAGKK604 pKa = 9.87IPAWLNYY611 pKa = 6.66MTDD614 pKa = 3.13VNEE617 pKa = 4.49TYY619 pKa = 11.45GNFADD624 pKa = 3.94NDD626 pKa = 3.9EE627 pKa = 4.81TSGQMWMTLNRR638 pKa = 11.84NYY640 pKa = 10.56EE641 pKa = 4.03RR642 pKa = 11.84TNDD645 pKa = 4.33GIGDD649 pKa = 4.18LTTYY653 pKa = 10.58IDD655 pKa = 3.63PTKK658 pKa = 10.63YY659 pKa = 10.59NYY661 pKa = 10.92AFAVKK666 pKa = 10.23DD667 pKa = 3.72LEE669 pKa = 4.43AQNFWAQIGMNIVARR684 pKa = 11.84RR685 pKa = 11.84KK686 pKa = 9.14MSAKK690 pKa = 10.1QIPNLL695 pKa = 3.74
Molecular weight: 79.39 kDa
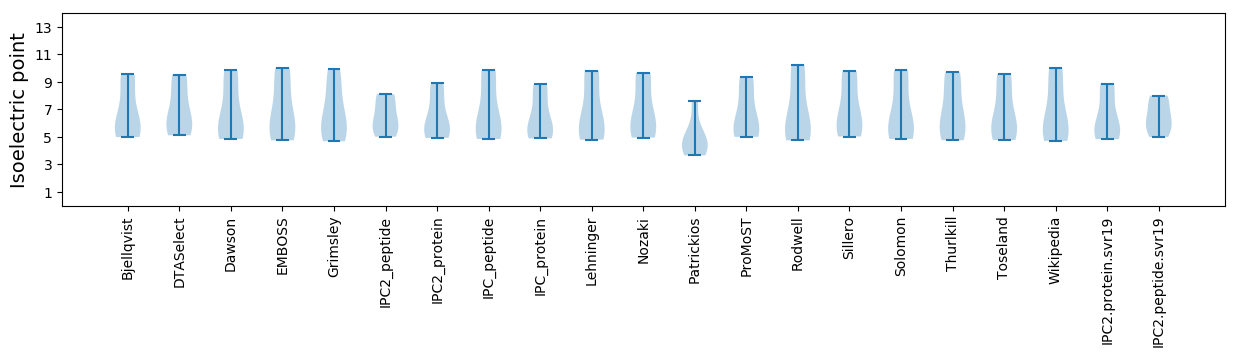
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W7L2|A0A4P8W7L2_9VIRU Uncharacterized protein OS=Capybara microvirus Cap1_SP_81 OX=2584792 PE=4 SV=1
MM1 pKa = 7.62CLYY4 pKa = 10.06PRR6 pKa = 11.84TIRR9 pKa = 11.84NPRR12 pKa = 11.84FAVGNKK18 pKa = 9.45NGYY21 pKa = 8.55SQDD24 pKa = 3.48DD25 pKa = 3.89CTDD28 pKa = 2.97KK29 pKa = 11.16RR30 pKa = 11.84KK31 pKa = 10.32LFIKK35 pKa = 9.8IDD37 pKa = 3.64CGNCEE42 pKa = 3.6EE43 pKa = 4.82CKK45 pKa = 10.06RR46 pKa = 11.84KK47 pKa = 9.67RR48 pKa = 11.84GKK50 pKa = 8.52WWTIRR55 pKa = 11.84IMEE58 pKa = 4.28EE59 pKa = 3.66LKK61 pKa = 10.7VNKK64 pKa = 8.83GTFVTLTFSAHH75 pKa = 6.83SLLQLTKK82 pKa = 10.9GEE84 pKa = 4.24KK85 pKa = 9.66TPTANEE91 pKa = 4.01TARR94 pKa = 11.84KK95 pKa = 8.41AVRR98 pKa = 11.84RR99 pKa = 11.84FLEE102 pKa = 4.38RR103 pKa = 11.84YY104 pKa = 7.03RR105 pKa = 11.84KK106 pKa = 10.06KK107 pKa = 9.62YY108 pKa = 9.05GHH110 pKa = 6.42SIKK113 pKa = 10.49HH114 pKa = 5.2ILITEE119 pKa = 4.63LGDD122 pKa = 4.64DD123 pKa = 4.22EE124 pKa = 5.51NEE126 pKa = 4.0HH127 pKa = 6.25TGRR130 pKa = 11.84IHH132 pKa = 6.54LHH134 pKa = 6.43GILFDD139 pKa = 4.58TITKK143 pKa = 10.59DD144 pKa = 3.03EE145 pKa = 5.37LYY147 pKa = 11.07NLWQYY152 pKa = 11.66GYY154 pKa = 10.33VDD156 pKa = 3.24TGSYY160 pKa = 10.45CNEE163 pKa = 3.41RR164 pKa = 11.84TANYY168 pKa = 5.37ITKK171 pKa = 10.37YY172 pKa = 8.01ITKK175 pKa = 9.91LDD177 pKa = 3.97TKK179 pKa = 10.59HH180 pKa = 6.83PEE182 pKa = 3.81YY183 pKa = 9.85KK184 pKa = 10.17QKK186 pKa = 10.74IFISAGLGKK195 pKa = 10.39NYY197 pKa = 10.25VEE199 pKa = 3.95TAKK202 pKa = 10.45RR203 pKa = 11.84QKK205 pKa = 10.47KK206 pKa = 9.02KK207 pKa = 10.66VYY209 pKa = 9.93RR210 pKa = 11.84FTNGTQFPLPTYY222 pKa = 9.76YY223 pKa = 10.46KK224 pKa = 9.96NKK226 pKa = 9.54VFTEE230 pKa = 4.22EE231 pKa = 4.18EE232 pKa = 4.3RR233 pKa = 11.84EE234 pKa = 4.15QMWTDD239 pKa = 3.37EE240 pKa = 4.28LNKK243 pKa = 10.2NEE245 pKa = 4.46HH246 pKa = 6.88YY247 pKa = 9.61ITGITYY253 pKa = 9.14RR254 pKa = 11.84YY255 pKa = 7.68RR256 pKa = 11.84AGVIGIKK263 pKa = 10.42GKK265 pKa = 8.67EE266 pKa = 3.79LEE268 pKa = 4.02QARR271 pKa = 11.84RR272 pKa = 11.84EE273 pKa = 4.14NEE275 pKa = 4.11TRR277 pKa = 11.84KK278 pKa = 9.41HH279 pKa = 4.86AQIANEE285 pKa = 3.81NKK287 pKa = 10.49GYY289 pKa = 11.3GNTKK293 pKa = 7.83KK294 pKa = 9.29TKK296 pKa = 10.19DD297 pKa = 3.31VLINEE302 pKa = 4.46KK303 pKa = 9.57MLQNSQKK310 pKa = 10.68EE311 pKa = 4.22EE312 pKa = 4.12QKK314 pKa = 11.33DD315 pKa = 3.53KK316 pKa = 11.67
MM1 pKa = 7.62CLYY4 pKa = 10.06PRR6 pKa = 11.84TIRR9 pKa = 11.84NPRR12 pKa = 11.84FAVGNKK18 pKa = 9.45NGYY21 pKa = 8.55SQDD24 pKa = 3.48DD25 pKa = 3.89CTDD28 pKa = 2.97KK29 pKa = 11.16RR30 pKa = 11.84KK31 pKa = 10.32LFIKK35 pKa = 9.8IDD37 pKa = 3.64CGNCEE42 pKa = 3.6EE43 pKa = 4.82CKK45 pKa = 10.06RR46 pKa = 11.84KK47 pKa = 9.67RR48 pKa = 11.84GKK50 pKa = 8.52WWTIRR55 pKa = 11.84IMEE58 pKa = 4.28EE59 pKa = 3.66LKK61 pKa = 10.7VNKK64 pKa = 8.83GTFVTLTFSAHH75 pKa = 6.83SLLQLTKK82 pKa = 10.9GEE84 pKa = 4.24KK85 pKa = 9.66TPTANEE91 pKa = 4.01TARR94 pKa = 11.84KK95 pKa = 8.41AVRR98 pKa = 11.84RR99 pKa = 11.84FLEE102 pKa = 4.38RR103 pKa = 11.84YY104 pKa = 7.03RR105 pKa = 11.84KK106 pKa = 10.06KK107 pKa = 9.62YY108 pKa = 9.05GHH110 pKa = 6.42SIKK113 pKa = 10.49HH114 pKa = 5.2ILITEE119 pKa = 4.63LGDD122 pKa = 4.64DD123 pKa = 4.22EE124 pKa = 5.51NEE126 pKa = 4.0HH127 pKa = 6.25TGRR130 pKa = 11.84IHH132 pKa = 6.54LHH134 pKa = 6.43GILFDD139 pKa = 4.58TITKK143 pKa = 10.59DD144 pKa = 3.03EE145 pKa = 5.37LYY147 pKa = 11.07NLWQYY152 pKa = 11.66GYY154 pKa = 10.33VDD156 pKa = 3.24TGSYY160 pKa = 10.45CNEE163 pKa = 3.41RR164 pKa = 11.84TANYY168 pKa = 5.37ITKK171 pKa = 10.37YY172 pKa = 8.01ITKK175 pKa = 9.91LDD177 pKa = 3.97TKK179 pKa = 10.59HH180 pKa = 6.83PEE182 pKa = 3.81YY183 pKa = 9.85KK184 pKa = 10.17QKK186 pKa = 10.74IFISAGLGKK195 pKa = 10.39NYY197 pKa = 10.25VEE199 pKa = 3.95TAKK202 pKa = 10.45RR203 pKa = 11.84QKK205 pKa = 10.47KK206 pKa = 9.02KK207 pKa = 10.66VYY209 pKa = 9.93RR210 pKa = 11.84FTNGTQFPLPTYY222 pKa = 9.76YY223 pKa = 10.46KK224 pKa = 9.96NKK226 pKa = 9.54VFTEE230 pKa = 4.22EE231 pKa = 4.18EE232 pKa = 4.3RR233 pKa = 11.84EE234 pKa = 4.15QMWTDD239 pKa = 3.37EE240 pKa = 4.28LNKK243 pKa = 10.2NEE245 pKa = 4.46HH246 pKa = 6.88YY247 pKa = 9.61ITGITYY253 pKa = 9.14RR254 pKa = 11.84YY255 pKa = 7.68RR256 pKa = 11.84AGVIGIKK263 pKa = 10.42GKK265 pKa = 8.67EE266 pKa = 3.79LEE268 pKa = 4.02QARR271 pKa = 11.84RR272 pKa = 11.84EE273 pKa = 4.14NEE275 pKa = 4.11TRR277 pKa = 11.84KK278 pKa = 9.41HH279 pKa = 4.86AQIANEE285 pKa = 3.81NKK287 pKa = 10.49GYY289 pKa = 11.3GNTKK293 pKa = 7.83KK294 pKa = 9.29TKK296 pKa = 10.19DD297 pKa = 3.31VLINEE302 pKa = 4.46KK303 pKa = 9.57MLQNSQKK310 pKa = 10.68EE311 pKa = 4.22EE312 pKa = 4.12QKK314 pKa = 11.33DD315 pKa = 3.53KK316 pKa = 11.67
Molecular weight: 37.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1622 |
75 |
695 |
270.3 |
30.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.597 ± 1.211 | 0.863 ± 0.411 |
5.117 ± 0.577 | 7.275 ± 1.004 |
3.268 ± 0.598 | 7.707 ± 1.484 |
1.665 ± 0.395 | 7.522 ± 1.342 |
7.707 ± 1.602 | 7.398 ± 0.651 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.466 ± 0.423 | 8.57 ± 0.491 |
2.713 ± 0.642 | 4.932 ± 0.577 |
4.254 ± 0.857 | 4.501 ± 0.929 |
7.213 ± 0.751 | 3.329 ± 0.348 |
1.85 ± 0.264 | 5.055 ± 0.669 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
