
Xingshan nematode virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
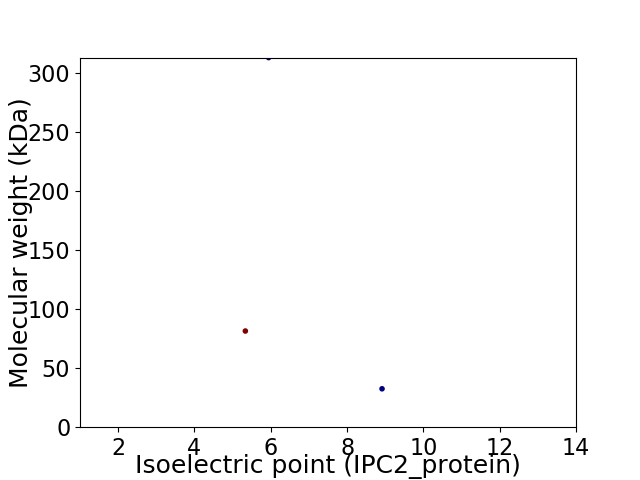
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KKI9|A0A1L3KKI9_9VIRU DiSB-ORF2_chro domain-containing protein OS=Xingshan nematode virus 2 OX=1923761 PE=4 SV=1
MM1 pKa = 7.56RR2 pKa = 11.84VCVLLISFFGAFLGSTEE19 pKa = 4.12RR20 pKa = 11.84FGLRR24 pKa = 11.84YY25 pKa = 9.53HH26 pKa = 6.07EE27 pKa = 4.71RR28 pKa = 11.84ARR30 pKa = 11.84KK31 pKa = 9.3IPGAYY36 pKa = 9.18DD37 pKa = 3.3VYY39 pKa = 11.1LDD41 pKa = 3.93SLISKK46 pKa = 9.69YY47 pKa = 10.87EE48 pKa = 3.76NASYY52 pKa = 10.39YY53 pKa = 10.88LKK55 pKa = 10.61IFGLVTSNQAKK66 pKa = 10.18RR67 pKa = 11.84LIEE70 pKa = 4.2EE71 pKa = 4.38LDD73 pKa = 3.68SLRR76 pKa = 11.84KK77 pKa = 9.5EE78 pKa = 3.76IQDD81 pKa = 3.28RR82 pKa = 11.84VDD84 pKa = 3.56YY85 pKa = 11.15VSEE88 pKa = 3.77NHH90 pKa = 6.22KK91 pKa = 10.25RR92 pKa = 11.84MSMIPFGVLANRR104 pKa = 11.84YY105 pKa = 8.61IVDD108 pKa = 3.8DD109 pKa = 4.01AVVAGFRR116 pKa = 11.84SIDD119 pKa = 3.39YY120 pKa = 9.93YY121 pKa = 11.27RR122 pKa = 11.84RR123 pKa = 11.84ILSFCLADD131 pKa = 3.62LEE133 pKa = 4.84SRR135 pKa = 11.84ILTVSEE141 pKa = 4.22TFLDD145 pKa = 4.45PVCMSAYY152 pKa = 9.74NKK154 pKa = 10.39SFNEE158 pKa = 5.03DD159 pKa = 2.66IDD161 pKa = 4.25CRR163 pKa = 11.84LSSLCFGVIKK173 pKa = 10.79KK174 pKa = 10.15NVDD177 pKa = 3.22GLFICSGYY185 pKa = 10.59SKK187 pKa = 11.12NLFRR191 pKa = 11.84HH192 pKa = 4.78YY193 pKa = 10.62RR194 pKa = 11.84LAKK197 pKa = 8.88STLLADD203 pKa = 3.72VFPLRR208 pKa = 11.84EE209 pKa = 4.15GQFEE213 pKa = 4.45FYY215 pKa = 10.97CNVSNNRR222 pKa = 11.84PLLVHH227 pKa = 6.98KK228 pKa = 10.75SLLNGDD234 pKa = 3.92KK235 pKa = 10.82RR236 pKa = 11.84FVDD239 pKa = 3.94PFSVLLQSDD248 pKa = 4.52FPFLTFSDD256 pKa = 3.33GGYY259 pKa = 9.28RR260 pKa = 11.84ALVFSQFVDD269 pKa = 3.55SYY271 pKa = 10.01STVSLATAGFKK282 pKa = 9.59YY283 pKa = 10.5VCNDD287 pKa = 3.18TLFAEE292 pKa = 4.7SSIIDD297 pKa = 3.76DD298 pKa = 3.73VNVFRR303 pKa = 11.84SFRR306 pKa = 11.84GCRR309 pKa = 11.84NFVLTEE315 pKa = 4.13GSDD318 pKa = 3.64VSSKK322 pKa = 10.43ICVDD326 pKa = 3.37HH327 pKa = 7.35LYY329 pKa = 10.79QDD331 pKa = 4.09FQALVDD337 pKa = 4.1RR338 pKa = 11.84CPEE341 pKa = 3.42DD342 pKa = 3.54WYY344 pKa = 10.17TFDD347 pKa = 3.85HH348 pKa = 6.77AVATFEE354 pKa = 4.61TYY356 pKa = 10.13TIGINDD362 pKa = 3.74YY363 pKa = 11.54SEE365 pKa = 5.04VLWKK369 pKa = 9.08TIPQVVTDD377 pKa = 4.01LLRR380 pKa = 11.84YY381 pKa = 9.52FDD383 pKa = 4.4RR384 pKa = 11.84NHH386 pKa = 6.1INVRR390 pKa = 11.84VQDD393 pKa = 3.62FEE395 pKa = 5.12KK396 pKa = 10.41FYY398 pKa = 11.22KK399 pKa = 10.86DD400 pKa = 3.06FVDD403 pKa = 3.89YY404 pKa = 10.98VRR406 pKa = 11.84SFGTLTNVTVRR417 pKa = 11.84FADD420 pKa = 3.44QVFDD424 pKa = 4.9GIGANLRR431 pKa = 11.84LLDD434 pKa = 3.84SFTFRR439 pKa = 11.84NFSEE443 pKa = 4.43NFVDD447 pKa = 4.45YY448 pKa = 10.02FWAKK452 pKa = 10.84AMGVDD457 pKa = 3.81VEE459 pKa = 4.38QIRR462 pKa = 11.84DD463 pKa = 3.65TTHH466 pKa = 6.44FEE468 pKa = 4.48SIHH471 pKa = 6.15SVTSVSGWLAQAISSFVRR489 pKa = 11.84PFWQVFLEE497 pKa = 4.01LLEE500 pKa = 5.64DD501 pKa = 3.45ILDD504 pKa = 3.96IIIRR508 pKa = 11.84VLFDD512 pKa = 3.52LTPLLEE518 pKa = 4.09KK519 pKa = 10.33FYY521 pKa = 10.85RR522 pKa = 11.84LTLAAFDD529 pKa = 4.19SLLNLLFQVLTLVLSFLLHH548 pKa = 6.4ILIILEE554 pKa = 4.23SKK556 pKa = 10.47ILLSEE561 pKa = 4.09YY562 pKa = 10.65LLVYY566 pKa = 10.4LFLSRR571 pKa = 11.84YY572 pKa = 7.19WSSPVPPLLILLLLILVFGFQRR594 pKa = 11.84SYY596 pKa = 11.05PSFLFFVLNHH606 pKa = 5.95QIKK609 pKa = 10.39SVLNATAIPSVTYY622 pKa = 10.14DD623 pKa = 3.17YY624 pKa = 11.5QFVYY628 pKa = 10.9NITLVNYY635 pKa = 7.34THH637 pKa = 6.99HH638 pKa = 7.42LYY640 pKa = 10.67HH641 pKa = 6.64FSIVNTPYY649 pKa = 10.21SYY651 pKa = 11.36SFYY654 pKa = 10.58VSNVSVFTYY663 pKa = 10.39VSEE666 pKa = 4.3KK667 pKa = 10.37FSSFNSTFFVTTFSHH682 pKa = 6.48YY683 pKa = 10.7LDD685 pKa = 4.86SSDD688 pKa = 5.38DD689 pKa = 3.72CDD691 pKa = 3.95FVCFLKK697 pKa = 11.0FFGFF701 pKa = 4.04
MM1 pKa = 7.56RR2 pKa = 11.84VCVLLISFFGAFLGSTEE19 pKa = 4.12RR20 pKa = 11.84FGLRR24 pKa = 11.84YY25 pKa = 9.53HH26 pKa = 6.07EE27 pKa = 4.71RR28 pKa = 11.84ARR30 pKa = 11.84KK31 pKa = 9.3IPGAYY36 pKa = 9.18DD37 pKa = 3.3VYY39 pKa = 11.1LDD41 pKa = 3.93SLISKK46 pKa = 9.69YY47 pKa = 10.87EE48 pKa = 3.76NASYY52 pKa = 10.39YY53 pKa = 10.88LKK55 pKa = 10.61IFGLVTSNQAKK66 pKa = 10.18RR67 pKa = 11.84LIEE70 pKa = 4.2EE71 pKa = 4.38LDD73 pKa = 3.68SLRR76 pKa = 11.84KK77 pKa = 9.5EE78 pKa = 3.76IQDD81 pKa = 3.28RR82 pKa = 11.84VDD84 pKa = 3.56YY85 pKa = 11.15VSEE88 pKa = 3.77NHH90 pKa = 6.22KK91 pKa = 10.25RR92 pKa = 11.84MSMIPFGVLANRR104 pKa = 11.84YY105 pKa = 8.61IVDD108 pKa = 3.8DD109 pKa = 4.01AVVAGFRR116 pKa = 11.84SIDD119 pKa = 3.39YY120 pKa = 9.93YY121 pKa = 11.27RR122 pKa = 11.84RR123 pKa = 11.84ILSFCLADD131 pKa = 3.62LEE133 pKa = 4.84SRR135 pKa = 11.84ILTVSEE141 pKa = 4.22TFLDD145 pKa = 4.45PVCMSAYY152 pKa = 9.74NKK154 pKa = 10.39SFNEE158 pKa = 5.03DD159 pKa = 2.66IDD161 pKa = 4.25CRR163 pKa = 11.84LSSLCFGVIKK173 pKa = 10.79KK174 pKa = 10.15NVDD177 pKa = 3.22GLFICSGYY185 pKa = 10.59SKK187 pKa = 11.12NLFRR191 pKa = 11.84HH192 pKa = 4.78YY193 pKa = 10.62RR194 pKa = 11.84LAKK197 pKa = 8.88STLLADD203 pKa = 3.72VFPLRR208 pKa = 11.84EE209 pKa = 4.15GQFEE213 pKa = 4.45FYY215 pKa = 10.97CNVSNNRR222 pKa = 11.84PLLVHH227 pKa = 6.98KK228 pKa = 10.75SLLNGDD234 pKa = 3.92KK235 pKa = 10.82RR236 pKa = 11.84FVDD239 pKa = 3.94PFSVLLQSDD248 pKa = 4.52FPFLTFSDD256 pKa = 3.33GGYY259 pKa = 9.28RR260 pKa = 11.84ALVFSQFVDD269 pKa = 3.55SYY271 pKa = 10.01STVSLATAGFKK282 pKa = 9.59YY283 pKa = 10.5VCNDD287 pKa = 3.18TLFAEE292 pKa = 4.7SSIIDD297 pKa = 3.76DD298 pKa = 3.73VNVFRR303 pKa = 11.84SFRR306 pKa = 11.84GCRR309 pKa = 11.84NFVLTEE315 pKa = 4.13GSDD318 pKa = 3.64VSSKK322 pKa = 10.43ICVDD326 pKa = 3.37HH327 pKa = 7.35LYY329 pKa = 10.79QDD331 pKa = 4.09FQALVDD337 pKa = 4.1RR338 pKa = 11.84CPEE341 pKa = 3.42DD342 pKa = 3.54WYY344 pKa = 10.17TFDD347 pKa = 3.85HH348 pKa = 6.77AVATFEE354 pKa = 4.61TYY356 pKa = 10.13TIGINDD362 pKa = 3.74YY363 pKa = 11.54SEE365 pKa = 5.04VLWKK369 pKa = 9.08TIPQVVTDD377 pKa = 4.01LLRR380 pKa = 11.84YY381 pKa = 9.52FDD383 pKa = 4.4RR384 pKa = 11.84NHH386 pKa = 6.1INVRR390 pKa = 11.84VQDD393 pKa = 3.62FEE395 pKa = 5.12KK396 pKa = 10.41FYY398 pKa = 11.22KK399 pKa = 10.86DD400 pKa = 3.06FVDD403 pKa = 3.89YY404 pKa = 10.98VRR406 pKa = 11.84SFGTLTNVTVRR417 pKa = 11.84FADD420 pKa = 3.44QVFDD424 pKa = 4.9GIGANLRR431 pKa = 11.84LLDD434 pKa = 3.84SFTFRR439 pKa = 11.84NFSEE443 pKa = 4.43NFVDD447 pKa = 4.45YY448 pKa = 10.02FWAKK452 pKa = 10.84AMGVDD457 pKa = 3.81VEE459 pKa = 4.38QIRR462 pKa = 11.84DD463 pKa = 3.65TTHH466 pKa = 6.44FEE468 pKa = 4.48SIHH471 pKa = 6.15SVTSVSGWLAQAISSFVRR489 pKa = 11.84PFWQVFLEE497 pKa = 4.01LLEE500 pKa = 5.64DD501 pKa = 3.45ILDD504 pKa = 3.96IIIRR508 pKa = 11.84VLFDD512 pKa = 3.52LTPLLEE518 pKa = 4.09KK519 pKa = 10.33FYY521 pKa = 10.85RR522 pKa = 11.84LTLAAFDD529 pKa = 4.19SLLNLLFQVLTLVLSFLLHH548 pKa = 6.4ILIILEE554 pKa = 4.23SKK556 pKa = 10.47ILLSEE561 pKa = 4.09YY562 pKa = 10.65LLVYY566 pKa = 10.4LFLSRR571 pKa = 11.84YY572 pKa = 7.19WSSPVPPLLILLLLILVFGFQRR594 pKa = 11.84SYY596 pKa = 11.05PSFLFFVLNHH606 pKa = 5.95QIKK609 pKa = 10.39SVLNATAIPSVTYY622 pKa = 10.14DD623 pKa = 3.17YY624 pKa = 11.5QFVYY628 pKa = 10.9NITLVNYY635 pKa = 7.34THH637 pKa = 6.99HH638 pKa = 7.42LYY640 pKa = 10.67HH641 pKa = 6.64FSIVNTPYY649 pKa = 10.21SYY651 pKa = 11.36SFYY654 pKa = 10.58VSNVSVFTYY663 pKa = 10.39VSEE666 pKa = 4.3KK667 pKa = 10.37FSSFNSTFFVTTFSHH682 pKa = 6.48YY683 pKa = 10.7LDD685 pKa = 4.86SSDD688 pKa = 5.38DD689 pKa = 3.72CDD691 pKa = 3.95FVCFLKK697 pKa = 11.0FFGFF701 pKa = 4.04
Molecular weight: 81.35 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KKI9|A0A1L3KKI9_9VIRU DiSB-ORF2_chro domain-containing protein OS=Xingshan nematode virus 2 OX=1923761 PE=4 SV=1
MM1 pKa = 7.54ASSALMLPSEE11 pKa = 4.49NGRR14 pKa = 11.84PPKK17 pKa = 10.36FYY19 pKa = 11.19DD20 pKa = 3.26HH21 pKa = 6.94ATGIQTVRR29 pKa = 11.84GGQQVGEE36 pKa = 4.45GFFSSVFNSFYY47 pKa = 10.86NIIYY51 pKa = 9.0HH52 pKa = 6.15FWGLTFAVLGIFTLLSEE69 pKa = 4.67YY70 pKa = 9.52GTASGPLEE78 pKa = 4.12VLLQAILKK86 pKa = 8.92FISDD90 pKa = 3.93PDD92 pKa = 3.62VPIVLKK98 pKa = 10.96SIASAFAWLLGYY110 pKa = 7.75MVKK113 pKa = 10.52YY114 pKa = 9.75KK115 pKa = 10.98YY116 pKa = 10.48VVAYY120 pKa = 9.9SAILFVPAIVKK131 pKa = 10.03PSTRR135 pKa = 11.84NIMFSFLLIFMAFLHH150 pKa = 6.66YY151 pKa = 9.58ITILQVLLLSLLFYY165 pKa = 10.45LFVMLRR171 pKa = 11.84TPAHH175 pKa = 6.14KK176 pKa = 10.4FFVLFMALIVFSVGFTEE193 pKa = 4.27HH194 pKa = 6.76QSFLSSKK201 pKa = 10.91LNLLLPADD209 pKa = 4.21LSKK212 pKa = 11.02PLTFKK217 pKa = 10.94PNTFLLPDD225 pKa = 3.9AQSPSARR232 pKa = 11.84AKK234 pKa = 10.1RR235 pKa = 11.84DD236 pKa = 3.2EE237 pKa = 4.1NEE239 pKa = 3.88ILRR242 pKa = 11.84HH243 pKa = 5.09QVAQLMEE250 pKa = 5.47KK251 pKa = 9.74ISNMHH256 pKa = 6.03QEE258 pKa = 4.26MEE260 pKa = 4.38TMRR263 pKa = 11.84QAASKK268 pKa = 10.13PNEE271 pKa = 4.11ATTAAPTVLPRR282 pKa = 11.84TEE284 pKa = 3.84RR285 pKa = 11.84HH286 pKa = 5.66KK287 pKa = 11.28SSS289 pKa = 3.12
MM1 pKa = 7.54ASSALMLPSEE11 pKa = 4.49NGRR14 pKa = 11.84PPKK17 pKa = 10.36FYY19 pKa = 11.19DD20 pKa = 3.26HH21 pKa = 6.94ATGIQTVRR29 pKa = 11.84GGQQVGEE36 pKa = 4.45GFFSSVFNSFYY47 pKa = 10.86NIIYY51 pKa = 9.0HH52 pKa = 6.15FWGLTFAVLGIFTLLSEE69 pKa = 4.67YY70 pKa = 9.52GTASGPLEE78 pKa = 4.12VLLQAILKK86 pKa = 8.92FISDD90 pKa = 3.93PDD92 pKa = 3.62VPIVLKK98 pKa = 10.96SIASAFAWLLGYY110 pKa = 7.75MVKK113 pKa = 10.52YY114 pKa = 9.75KK115 pKa = 10.98YY116 pKa = 10.48VVAYY120 pKa = 9.9SAILFVPAIVKK131 pKa = 10.03PSTRR135 pKa = 11.84NIMFSFLLIFMAFLHH150 pKa = 6.66YY151 pKa = 9.58ITILQVLLLSLLFYY165 pKa = 10.45LFVMLRR171 pKa = 11.84TPAHH175 pKa = 6.14KK176 pKa = 10.4FFVLFMALIVFSVGFTEE193 pKa = 4.27HH194 pKa = 6.76QSFLSSKK201 pKa = 10.91LNLLLPADD209 pKa = 4.21LSKK212 pKa = 11.02PLTFKK217 pKa = 10.94PNTFLLPDD225 pKa = 3.9AQSPSARR232 pKa = 11.84AKK234 pKa = 10.1RR235 pKa = 11.84DD236 pKa = 3.2EE237 pKa = 4.1NEE239 pKa = 3.88ILRR242 pKa = 11.84HH243 pKa = 5.09QVAQLMEE250 pKa = 5.47KK251 pKa = 9.74ISNMHH256 pKa = 6.03QEE258 pKa = 4.26MEE260 pKa = 4.38TMRR263 pKa = 11.84QAASKK268 pKa = 10.13PNEE271 pKa = 4.11ATTAAPTVLPRR282 pKa = 11.84TEE284 pKa = 3.84RR285 pKa = 11.84HH286 pKa = 5.66KK287 pKa = 11.28SSS289 pKa = 3.12
Molecular weight: 32.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3712 |
289 |
2722 |
1237.3 |
142.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.55 ± 0.783 | 2.182 ± 0.574 |
7.462 ± 1.359 | 5.038 ± 0.559 |
6.897 ± 1.484 | 4.31 ± 0.301 |
1.913 ± 0.246 | 5.711 ± 0.06 |
6.008 ± 1.113 | 9.833 ± 1.511 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.586 ± 0.696 | 4.095 ± 0.225 |
2.963 ± 0.67 | 2.344 ± 0.362 |
5.792 ± 0.618 | 7.678 ± 0.926 |
4.984 ± 0.134 | 8.944 ± 0.546 |
0.7 ± 0.06 | 5.011 ± 0.39 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
