
Phaeodactylibacter luteus
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Saprospiria; Saprospirales; Haliscomenobacteraceae; Phaeodactylibacter
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
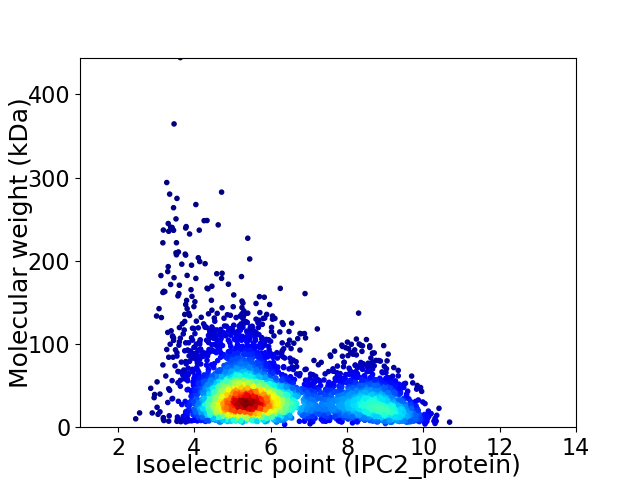
Virtual 2D-PAGE plot for 4366 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5C6RW63|A0A5C6RW63_9BACT T9SS type B sorting domain-containing protein OS=Phaeodactylibacter luteus OX=1564516 GN=FRY97_05585 PE=4 SV=1
MM1 pKa = 8.01DD2 pKa = 4.93KK3 pKa = 11.16ALATGMLLLHH13 pKa = 6.77SIFLSAQFAPPFDD26 pKa = 4.65LFDD29 pKa = 3.79EE30 pKa = 4.69VGDD33 pKa = 3.8VQFSIGSDD41 pKa = 3.06VDD43 pKa = 3.38GDD45 pKa = 3.55GRR47 pKa = 11.84TDD49 pKa = 3.84VICATAKK56 pKa = 10.38RR57 pKa = 11.84LFWLPSQADD66 pKa = 3.73GTFGHH71 pKa = 5.99QRR73 pKa = 11.84PISFADD79 pKa = 3.07QLYY82 pKa = 10.3RR83 pKa = 11.84SLTAFDD89 pKa = 3.96IDD91 pKa = 4.2ADD93 pKa = 3.92GDD95 pKa = 3.95EE96 pKa = 4.91DD97 pKa = 5.88LLLASYY103 pKa = 11.27NEE105 pKa = 4.76GIGIHH110 pKa = 6.17FNNGDD115 pKa = 3.62GTFGAVVIIYY125 pKa = 10.47SPGPVLDD132 pKa = 5.69LYY134 pKa = 11.45LADD137 pKa = 5.06LNGDD141 pKa = 4.13SLPDD145 pKa = 3.53ALVGQGQGVAWLPGNGDD162 pKa = 3.66GTFGAPSLIAAGHH175 pKa = 6.48DD176 pKa = 3.41AANSVQAEE184 pKa = 4.23DD185 pKa = 4.27LNGDD189 pKa = 3.56GKK191 pKa = 11.31LDD193 pKa = 3.64VVVAAHH199 pKa = 6.46SDD201 pKa = 3.45NRR203 pKa = 11.84LSWHH207 pKa = 6.53EE208 pKa = 3.96NLGDD212 pKa = 3.76GTFADD217 pKa = 3.85YY218 pKa = 11.05QIINEE223 pKa = 4.25THH225 pKa = 7.11LGPNSLDD232 pKa = 3.21IGDD235 pKa = 4.4FDD237 pKa = 5.66GDD239 pKa = 3.49GWPDD243 pKa = 3.32IVCSSRR249 pKa = 11.84NEE251 pKa = 4.1DD252 pKa = 3.81DD253 pKa = 4.28GGAGLALFRR262 pKa = 11.84NLGHH266 pKa = 6.97GSFAAPDD273 pKa = 3.99TLLPAYY279 pKa = 10.27GNYY282 pKa = 9.07FVQAGDD288 pKa = 3.45MDD290 pKa = 4.77GDD292 pKa = 3.98GDD294 pKa = 4.71LDD296 pKa = 3.72LVSCHH301 pKa = 6.99RR302 pKa = 11.84YY303 pKa = 10.57GEE305 pKa = 4.42VGWFQNDD312 pKa = 3.53GTGHH316 pKa = 7.13FGPDD320 pKa = 3.46IKK322 pKa = 11.09LQDD325 pKa = 3.72YY326 pKa = 9.37AAHH329 pKa = 6.23QRR331 pKa = 11.84FLDD334 pKa = 4.15LPDD337 pKa = 4.43LNGDD341 pKa = 3.92GLPDD345 pKa = 3.19ILYY348 pKa = 10.43GINRR352 pKa = 11.84PNPPKK357 pKa = 10.42EE358 pKa = 3.79ALLWLASEE366 pKa = 4.8GGGTFSPPALLEE378 pKa = 4.38LGVSQVSIPVVADD391 pKa = 3.2MDD393 pKa = 3.81QDD395 pKa = 3.52GRR397 pKa = 11.84PDD399 pKa = 3.42VVLGSSGSDD408 pKa = 2.92ALIWVRR414 pKa = 11.84NEE416 pKa = 3.28GHH418 pKa = 6.83GYY420 pKa = 8.39SQPRR424 pKa = 11.84LIARR428 pKa = 11.84RR429 pKa = 11.84HH430 pKa = 5.22NYY432 pKa = 9.06EE433 pKa = 3.85PASIAVFDD441 pKa = 4.64FNNDD445 pKa = 2.5GWPDD449 pKa = 3.48VLAGSVADD457 pKa = 4.24DD458 pKa = 3.83ATNQSMFCVYY468 pKa = 10.67LNNADD473 pKa = 3.57GTFAATPPFEE483 pKa = 4.85SGYY486 pKa = 10.19FYY488 pKa = 11.16KK489 pKa = 10.24RR490 pKa = 11.84DD491 pKa = 3.39LQMADD496 pKa = 3.0MDD498 pKa = 4.66GDD500 pKa = 4.16GDD502 pKa = 6.54LDD504 pKa = 4.08MLWASPGFNGQPTGYY519 pKa = 10.49VGWARR524 pKa = 11.84NNGDD528 pKa = 2.55SWGNFVYY535 pKa = 10.75LLEE538 pKa = 4.43EE539 pKa = 4.12AVGVLDD545 pKa = 5.47VLAADD550 pKa = 4.87LNGDD554 pKa = 3.96GLNDD558 pKa = 3.67LVFCQGEE565 pKa = 4.6SPAIDD570 pKa = 3.22YY571 pKa = 10.34RR572 pKa = 11.84LNQGNGNFVSLQTIANDD589 pKa = 3.33FTTALYY595 pKa = 10.33AQAADD600 pKa = 4.06LNGDD604 pKa = 3.55GLMDD608 pKa = 5.67IIATADD614 pKa = 3.26NSFGGNDD621 pKa = 3.88QIAWWPGTGGGFFGPQQPIYY641 pKa = 10.12IGGPAMEE648 pKa = 5.07KK649 pKa = 10.35FALTDD654 pKa = 3.56YY655 pKa = 11.51NNDD658 pKa = 3.44GEE660 pKa = 4.8LDD662 pKa = 4.03IITSGYY668 pKa = 10.36QDD670 pKa = 5.73FPMQWLEE677 pKa = 3.98GQGGGSFLPPLPIPNAAIAVRR698 pKa = 11.84GFEE701 pKa = 5.58LADD704 pKa = 4.05LDD706 pKa = 4.52NDD708 pKa = 3.77GDD710 pKa = 4.24LDD712 pKa = 3.68IVGNAEE718 pKa = 3.99SPPNGGIDD726 pKa = 3.58RR727 pKa = 11.84AIISYY732 pKa = 10.74NLANDD737 pKa = 3.55VGVSGAVFFDD747 pKa = 3.91EE748 pKa = 4.46NANGLRR754 pKa = 11.84DD755 pKa = 3.62TSEE758 pKa = 4.02YY759 pKa = 11.08LLDD762 pKa = 3.97RR763 pKa = 11.84FPITVAPEE771 pKa = 3.67ALAVFSDD778 pKa = 3.69SEE780 pKa = 4.53GRR782 pKa = 11.84FSVYY786 pKa = 10.1GAEE789 pKa = 4.32GTYY792 pKa = 10.39SISPQLDD799 pKa = 3.77DD800 pKa = 4.18CWSLTTMPGSYY811 pKa = 10.2EE812 pKa = 3.84IVFDD816 pKa = 3.78GTPVDD821 pKa = 3.55TLLFGVSPNTEE832 pKa = 4.01TPEE835 pKa = 4.26GSVTLASAPTRR846 pKa = 11.84CGFTVPFWLNFNNDD860 pKa = 3.37GCWPFDD866 pKa = 3.47GQAFLALNEE875 pKa = 3.97LATYY879 pKa = 10.23IDD881 pKa = 4.59ASPAPSSVSGDD892 pKa = 3.39TLFWSFSALQPGEE905 pKa = 4.13SRR907 pKa = 11.84SINLNMEE914 pKa = 3.78IAGVEE919 pKa = 4.65FIGSALDD926 pKa = 3.76LPSGLVPYY934 pKa = 9.56NAQGEE939 pKa = 4.32ALPAEE944 pKa = 4.56VFAFTSIINCAYY956 pKa = 10.57DD957 pKa = 3.87PNDD960 pKa = 3.82KK961 pKa = 10.25LAHH964 pKa = 6.51PARR967 pKa = 11.84AEE969 pKa = 3.87QPPFSQNYY977 pKa = 7.41TLFDD981 pKa = 3.43EE982 pKa = 4.65PLRR985 pKa = 11.84YY986 pKa = 8.31TIRR989 pKa = 11.84FQNTGTDD996 pKa = 3.12TAFNVVLRR1004 pKa = 11.84DD1005 pKa = 3.67QLSEE1009 pKa = 3.96EE1010 pKa = 4.62LNWEE1014 pKa = 4.05TFEE1017 pKa = 5.12PGASSHH1023 pKa = 6.68PYY1025 pKa = 9.79EE1026 pKa = 4.32ATLYY1030 pKa = 10.93DD1031 pKa = 4.26DD1032 pKa = 4.39GQAAFHH1038 pKa = 6.37FRR1040 pKa = 11.84NILLPDD1046 pKa = 3.41STTNEE1051 pKa = 4.26PGSHH1055 pKa = 6.11GFVQFEE1061 pKa = 4.2IEE1063 pKa = 4.39ALPGLDD1069 pKa = 3.28EE1070 pKa = 4.28NTPIEE1075 pKa = 4.38NTAGIYY1081 pKa = 10.26FDD1083 pKa = 4.95FNPPIITNTVTNLMVSALPDD1103 pKa = 3.74STPVAAFGYY1112 pKa = 9.65EE1113 pKa = 3.99AFSLLVQFNDD1123 pKa = 4.02ASGLSPDD1130 pKa = 2.73SWFWDD1135 pKa = 4.1FGDD1138 pKa = 4.68GGSSTLQHH1146 pKa = 6.97PEE1148 pKa = 3.39HH1149 pKa = 6.89TYY1151 pKa = 11.26ASPGSYY1157 pKa = 9.17EE1158 pKa = 4.14VCLTAANAWGTAQTCQEE1175 pKa = 4.16VTLSSTGTVSVAPVALPEE1193 pKa = 4.23VFPNPARR1200 pKa = 11.84DD1201 pKa = 4.01KK1202 pKa = 11.23VWVKK1206 pKa = 8.67YY1207 pKa = 10.14QPLPSTAPLKK1217 pKa = 10.51IYY1219 pKa = 10.14NAYY1222 pKa = 8.85GQLVLTLPLPSGRR1235 pKa = 11.84SSWPTAGWPAGQYY1248 pKa = 8.51WLQTPSGEE1256 pKa = 3.73RR1257 pKa = 11.84VPFIILKK1264 pKa = 10.22
MM1 pKa = 8.01DD2 pKa = 4.93KK3 pKa = 11.16ALATGMLLLHH13 pKa = 6.77SIFLSAQFAPPFDD26 pKa = 4.65LFDD29 pKa = 3.79EE30 pKa = 4.69VGDD33 pKa = 3.8VQFSIGSDD41 pKa = 3.06VDD43 pKa = 3.38GDD45 pKa = 3.55GRR47 pKa = 11.84TDD49 pKa = 3.84VICATAKK56 pKa = 10.38RR57 pKa = 11.84LFWLPSQADD66 pKa = 3.73GTFGHH71 pKa = 5.99QRR73 pKa = 11.84PISFADD79 pKa = 3.07QLYY82 pKa = 10.3RR83 pKa = 11.84SLTAFDD89 pKa = 3.96IDD91 pKa = 4.2ADD93 pKa = 3.92GDD95 pKa = 3.95EE96 pKa = 4.91DD97 pKa = 5.88LLLASYY103 pKa = 11.27NEE105 pKa = 4.76GIGIHH110 pKa = 6.17FNNGDD115 pKa = 3.62GTFGAVVIIYY125 pKa = 10.47SPGPVLDD132 pKa = 5.69LYY134 pKa = 11.45LADD137 pKa = 5.06LNGDD141 pKa = 4.13SLPDD145 pKa = 3.53ALVGQGQGVAWLPGNGDD162 pKa = 3.66GTFGAPSLIAAGHH175 pKa = 6.48DD176 pKa = 3.41AANSVQAEE184 pKa = 4.23DD185 pKa = 4.27LNGDD189 pKa = 3.56GKK191 pKa = 11.31LDD193 pKa = 3.64VVVAAHH199 pKa = 6.46SDD201 pKa = 3.45NRR203 pKa = 11.84LSWHH207 pKa = 6.53EE208 pKa = 3.96NLGDD212 pKa = 3.76GTFADD217 pKa = 3.85YY218 pKa = 11.05QIINEE223 pKa = 4.25THH225 pKa = 7.11LGPNSLDD232 pKa = 3.21IGDD235 pKa = 4.4FDD237 pKa = 5.66GDD239 pKa = 3.49GWPDD243 pKa = 3.32IVCSSRR249 pKa = 11.84NEE251 pKa = 4.1DD252 pKa = 3.81DD253 pKa = 4.28GGAGLALFRR262 pKa = 11.84NLGHH266 pKa = 6.97GSFAAPDD273 pKa = 3.99TLLPAYY279 pKa = 10.27GNYY282 pKa = 9.07FVQAGDD288 pKa = 3.45MDD290 pKa = 4.77GDD292 pKa = 3.98GDD294 pKa = 4.71LDD296 pKa = 3.72LVSCHH301 pKa = 6.99RR302 pKa = 11.84YY303 pKa = 10.57GEE305 pKa = 4.42VGWFQNDD312 pKa = 3.53GTGHH316 pKa = 7.13FGPDD320 pKa = 3.46IKK322 pKa = 11.09LQDD325 pKa = 3.72YY326 pKa = 9.37AAHH329 pKa = 6.23QRR331 pKa = 11.84FLDD334 pKa = 4.15LPDD337 pKa = 4.43LNGDD341 pKa = 3.92GLPDD345 pKa = 3.19ILYY348 pKa = 10.43GINRR352 pKa = 11.84PNPPKK357 pKa = 10.42EE358 pKa = 3.79ALLWLASEE366 pKa = 4.8GGGTFSPPALLEE378 pKa = 4.38LGVSQVSIPVVADD391 pKa = 3.2MDD393 pKa = 3.81QDD395 pKa = 3.52GRR397 pKa = 11.84PDD399 pKa = 3.42VVLGSSGSDD408 pKa = 2.92ALIWVRR414 pKa = 11.84NEE416 pKa = 3.28GHH418 pKa = 6.83GYY420 pKa = 8.39SQPRR424 pKa = 11.84LIARR428 pKa = 11.84RR429 pKa = 11.84HH430 pKa = 5.22NYY432 pKa = 9.06EE433 pKa = 3.85PASIAVFDD441 pKa = 4.64FNNDD445 pKa = 2.5GWPDD449 pKa = 3.48VLAGSVADD457 pKa = 4.24DD458 pKa = 3.83ATNQSMFCVYY468 pKa = 10.67LNNADD473 pKa = 3.57GTFAATPPFEE483 pKa = 4.85SGYY486 pKa = 10.19FYY488 pKa = 11.16KK489 pKa = 10.24RR490 pKa = 11.84DD491 pKa = 3.39LQMADD496 pKa = 3.0MDD498 pKa = 4.66GDD500 pKa = 4.16GDD502 pKa = 6.54LDD504 pKa = 4.08MLWASPGFNGQPTGYY519 pKa = 10.49VGWARR524 pKa = 11.84NNGDD528 pKa = 2.55SWGNFVYY535 pKa = 10.75LLEE538 pKa = 4.43EE539 pKa = 4.12AVGVLDD545 pKa = 5.47VLAADD550 pKa = 4.87LNGDD554 pKa = 3.96GLNDD558 pKa = 3.67LVFCQGEE565 pKa = 4.6SPAIDD570 pKa = 3.22YY571 pKa = 10.34RR572 pKa = 11.84LNQGNGNFVSLQTIANDD589 pKa = 3.33FTTALYY595 pKa = 10.33AQAADD600 pKa = 4.06LNGDD604 pKa = 3.55GLMDD608 pKa = 5.67IIATADD614 pKa = 3.26NSFGGNDD621 pKa = 3.88QIAWWPGTGGGFFGPQQPIYY641 pKa = 10.12IGGPAMEE648 pKa = 5.07KK649 pKa = 10.35FALTDD654 pKa = 3.56YY655 pKa = 11.51NNDD658 pKa = 3.44GEE660 pKa = 4.8LDD662 pKa = 4.03IITSGYY668 pKa = 10.36QDD670 pKa = 5.73FPMQWLEE677 pKa = 3.98GQGGGSFLPPLPIPNAAIAVRR698 pKa = 11.84GFEE701 pKa = 5.58LADD704 pKa = 4.05LDD706 pKa = 4.52NDD708 pKa = 3.77GDD710 pKa = 4.24LDD712 pKa = 3.68IVGNAEE718 pKa = 3.99SPPNGGIDD726 pKa = 3.58RR727 pKa = 11.84AIISYY732 pKa = 10.74NLANDD737 pKa = 3.55VGVSGAVFFDD747 pKa = 3.91EE748 pKa = 4.46NANGLRR754 pKa = 11.84DD755 pKa = 3.62TSEE758 pKa = 4.02YY759 pKa = 11.08LLDD762 pKa = 3.97RR763 pKa = 11.84FPITVAPEE771 pKa = 3.67ALAVFSDD778 pKa = 3.69SEE780 pKa = 4.53GRR782 pKa = 11.84FSVYY786 pKa = 10.1GAEE789 pKa = 4.32GTYY792 pKa = 10.39SISPQLDD799 pKa = 3.77DD800 pKa = 4.18CWSLTTMPGSYY811 pKa = 10.2EE812 pKa = 3.84IVFDD816 pKa = 3.78GTPVDD821 pKa = 3.55TLLFGVSPNTEE832 pKa = 4.01TPEE835 pKa = 4.26GSVTLASAPTRR846 pKa = 11.84CGFTVPFWLNFNNDD860 pKa = 3.37GCWPFDD866 pKa = 3.47GQAFLALNEE875 pKa = 3.97LATYY879 pKa = 10.23IDD881 pKa = 4.59ASPAPSSVSGDD892 pKa = 3.39TLFWSFSALQPGEE905 pKa = 4.13SRR907 pKa = 11.84SINLNMEE914 pKa = 3.78IAGVEE919 pKa = 4.65FIGSALDD926 pKa = 3.76LPSGLVPYY934 pKa = 9.56NAQGEE939 pKa = 4.32ALPAEE944 pKa = 4.56VFAFTSIINCAYY956 pKa = 10.57DD957 pKa = 3.87PNDD960 pKa = 3.82KK961 pKa = 10.25LAHH964 pKa = 6.51PARR967 pKa = 11.84AEE969 pKa = 3.87QPPFSQNYY977 pKa = 7.41TLFDD981 pKa = 3.43EE982 pKa = 4.65PLRR985 pKa = 11.84YY986 pKa = 8.31TIRR989 pKa = 11.84FQNTGTDD996 pKa = 3.12TAFNVVLRR1004 pKa = 11.84DD1005 pKa = 3.67QLSEE1009 pKa = 3.96EE1010 pKa = 4.62LNWEE1014 pKa = 4.05TFEE1017 pKa = 5.12PGASSHH1023 pKa = 6.68PYY1025 pKa = 9.79EE1026 pKa = 4.32ATLYY1030 pKa = 10.93DD1031 pKa = 4.26DD1032 pKa = 4.39GQAAFHH1038 pKa = 6.37FRR1040 pKa = 11.84NILLPDD1046 pKa = 3.41STTNEE1051 pKa = 4.26PGSHH1055 pKa = 6.11GFVQFEE1061 pKa = 4.2IEE1063 pKa = 4.39ALPGLDD1069 pKa = 3.28EE1070 pKa = 4.28NTPIEE1075 pKa = 4.38NTAGIYY1081 pKa = 10.26FDD1083 pKa = 4.95FNPPIITNTVTNLMVSALPDD1103 pKa = 3.74STPVAAFGYY1112 pKa = 9.65EE1113 pKa = 3.99AFSLLVQFNDD1123 pKa = 4.02ASGLSPDD1130 pKa = 2.73SWFWDD1135 pKa = 4.1FGDD1138 pKa = 4.68GGSSTLQHH1146 pKa = 6.97PEE1148 pKa = 3.39HH1149 pKa = 6.89TYY1151 pKa = 11.26ASPGSYY1157 pKa = 9.17EE1158 pKa = 4.14VCLTAANAWGTAQTCQEE1175 pKa = 4.16VTLSSTGTVSVAPVALPEE1193 pKa = 4.23VFPNPARR1200 pKa = 11.84DD1201 pKa = 4.01KK1202 pKa = 11.23VWVKK1206 pKa = 8.67YY1207 pKa = 10.14QPLPSTAPLKK1217 pKa = 10.51IYY1219 pKa = 10.14NAYY1222 pKa = 8.85GQLVLTLPLPSGRR1235 pKa = 11.84SSWPTAGWPAGQYY1248 pKa = 8.51WLQTPSGEE1256 pKa = 3.73RR1257 pKa = 11.84VPFIILKK1264 pKa = 10.22
Molecular weight: 135.58 kDa
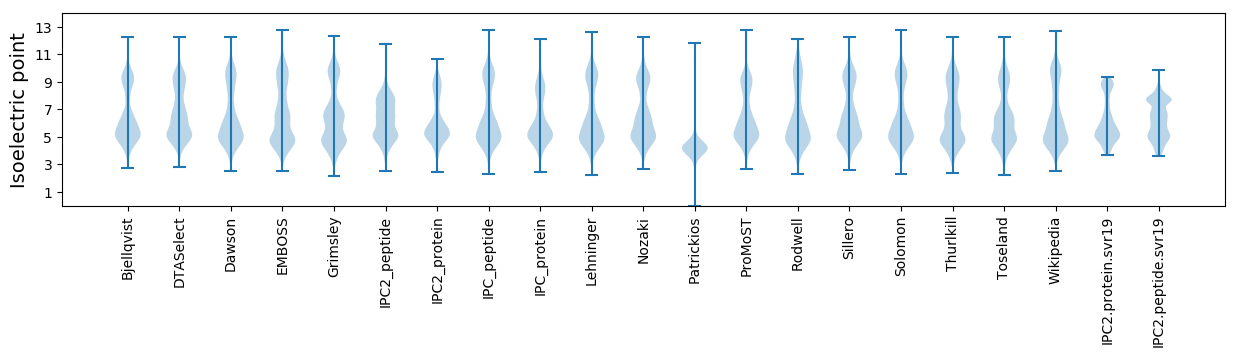
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5C6RL56|A0A5C6RL56_9BACT Uncharacterized protein OS=Phaeodactylibacter luteus OX=1564516 GN=FRY97_14410 PE=4 SV=1
MM1 pKa = 7.36MGPAPAARR9 pKa = 11.84AGAAMLRR16 pKa = 11.84GSLPARR22 pKa = 11.84PLALQASGLAFGHH35 pKa = 6.72PCAALGQCQDD45 pKa = 4.03IKK47 pKa = 10.12PQKK50 pKa = 9.5HH51 pKa = 3.54KK52 pKa = 10.13HH53 pKa = 4.73LRR55 pKa = 11.84YY56 pKa = 9.47FGSRR60 pKa = 11.84SGKK63 pKa = 9.88ALCC66 pKa = 4.94
MM1 pKa = 7.36MGPAPAARR9 pKa = 11.84AGAAMLRR16 pKa = 11.84GSLPARR22 pKa = 11.84PLALQASGLAFGHH35 pKa = 6.72PCAALGQCQDD45 pKa = 4.03IKK47 pKa = 10.12PQKK50 pKa = 9.5HH51 pKa = 3.54KK52 pKa = 10.13HH53 pKa = 4.73LRR55 pKa = 11.84YY56 pKa = 9.47FGSRR60 pKa = 11.84SGKK63 pKa = 9.88ALCC66 pKa = 4.94
Molecular weight: 6.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1681301 |
28 |
4203 |
385.1 |
42.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.532 ± 0.041 | 0.967 ± 0.023 |
5.25 ± 0.037 | 6.55 ± 0.043 |
4.649 ± 0.026 | 8.09 ± 0.049 |
1.902 ± 0.021 | 5.438 ± 0.03 |
4.223 ± 0.051 | 10.261 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.122 ± 0.02 | 4.026 ± 0.034 |
4.847 ± 0.034 | 4.509 ± 0.03 |
5.114 ± 0.045 | 6.06 ± 0.033 |
5.147 ± 0.046 | 6.238 ± 0.036 |
1.365 ± 0.015 | 3.711 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
