
Beihai barnacle virus 7
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.56
Get precalculated fractions of proteins
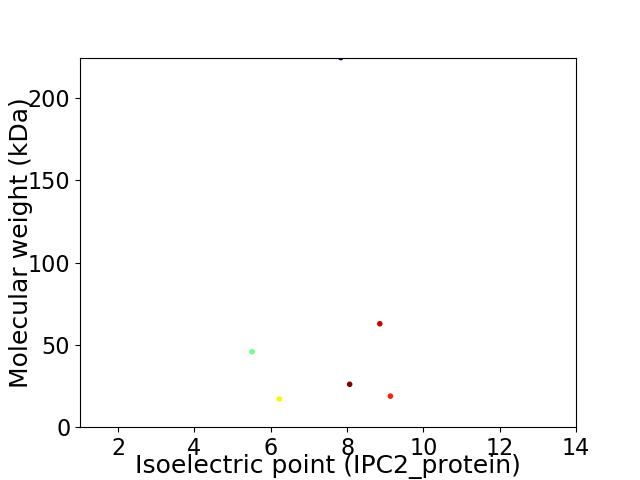
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KMQ3|A0A1L3KMQ3_9VIRU Putative glycoprotein OS=Beihai barnacle virus 7 OX=1922365 PE=4 SV=1
MM1 pKa = 6.37TTIKK5 pKa = 10.71AFLSGQVEE13 pKa = 4.21GLPASEE19 pKa = 4.24QEE21 pKa = 4.17PSNVEE26 pKa = 3.99CKK28 pKa = 10.41DD29 pKa = 3.41ALTDD33 pKa = 3.9RR34 pKa = 11.84LKK36 pKa = 10.81KK37 pKa = 10.12VYY39 pKa = 10.33NYY41 pKa = 10.53RR42 pKa = 11.84DD43 pKa = 3.88DD44 pKa = 3.45SHH46 pKa = 6.94KK47 pKa = 11.27ALLAKK52 pKa = 10.16AWRR55 pKa = 11.84EE56 pKa = 4.0RR57 pKa = 11.84NSDD60 pKa = 3.16KK61 pKa = 11.23ARR63 pKa = 11.84LVSRR67 pKa = 11.84KK68 pKa = 9.49LLLAAPLFYY77 pKa = 10.69SPPGSEE83 pKa = 3.41TWVRR87 pKa = 11.84TTAVKK92 pKa = 10.06PDD94 pKa = 3.54NASDD98 pKa = 4.45LIEE101 pKa = 5.38DD102 pKa = 3.94VLEE105 pKa = 4.57DD106 pKa = 3.83TLKK109 pKa = 10.18TDD111 pKa = 3.15ATTTPLTTLSQRR123 pKa = 11.84VLKK126 pKa = 10.61ILDD129 pKa = 3.54SSAGDD134 pKa = 3.39SLVSQEE140 pKa = 4.84TILEE144 pKa = 4.08YY145 pKa = 10.99LPIYY149 pKa = 10.3AIIPWRR155 pKa = 11.84ALVKK159 pKa = 10.65SGGVTSSEE167 pKa = 4.14WFSSATGAMTTAIKK181 pKa = 10.54AADD184 pKa = 3.71DD185 pKa = 3.8RR186 pKa = 11.84LRR188 pKa = 11.84QADD191 pKa = 3.88EE192 pKa = 4.1TVKK195 pKa = 11.1NDD197 pKa = 3.03MKK199 pKa = 10.71TWFAQVDD206 pKa = 3.89PTEE209 pKa = 4.58LLTNHH214 pKa = 6.98LAEE217 pKa = 4.94IMACALAHH225 pKa = 5.59SEE227 pKa = 4.11RR228 pKa = 11.84VPPFIAASIQSVWMYY243 pKa = 10.48HH244 pKa = 5.79GLGTYY249 pKa = 10.03IFLRR253 pKa = 11.84EE254 pKa = 3.91AARR257 pKa = 11.84TLGLNSPEE265 pKa = 4.29LLRR268 pKa = 11.84EE269 pKa = 4.33LCCPLLNNQIMALVKK284 pKa = 10.73LMIDD288 pKa = 3.37EE289 pKa = 4.36QANNTPVFPYY299 pKa = 10.35IKK301 pKa = 9.8IINTNFHH308 pKa = 6.49SGVSTKK314 pKa = 10.51RR315 pKa = 11.84CIPLMYY321 pKa = 10.13VLRR324 pKa = 11.84QIICPISPKK333 pKa = 9.36TKK335 pKa = 9.49QGEE338 pKa = 4.71GIWGKK343 pKa = 8.78MCGVPSLDD351 pKa = 3.17VRR353 pKa = 11.84IPLAQAAMRR362 pKa = 11.84LRR364 pKa = 11.84RR365 pKa = 11.84KK366 pKa = 10.03HH367 pKa = 5.74ILTVRR372 pKa = 11.84EE373 pKa = 4.48GEE375 pKa = 4.24TSEE378 pKa = 4.78VMQQILGEE386 pKa = 4.16IRR388 pKa = 11.84DD389 pKa = 4.08TPLGYY394 pKa = 10.4DD395 pKa = 3.51DD396 pKa = 4.46TSSEE400 pKa = 4.1QEE402 pKa = 4.14AVDD405 pKa = 4.78PDD407 pKa = 4.41DD408 pKa = 5.65LGNPPAA414 pKa = 5.56
MM1 pKa = 6.37TTIKK5 pKa = 10.71AFLSGQVEE13 pKa = 4.21GLPASEE19 pKa = 4.24QEE21 pKa = 4.17PSNVEE26 pKa = 3.99CKK28 pKa = 10.41DD29 pKa = 3.41ALTDD33 pKa = 3.9RR34 pKa = 11.84LKK36 pKa = 10.81KK37 pKa = 10.12VYY39 pKa = 10.33NYY41 pKa = 10.53RR42 pKa = 11.84DD43 pKa = 3.88DD44 pKa = 3.45SHH46 pKa = 6.94KK47 pKa = 11.27ALLAKK52 pKa = 10.16AWRR55 pKa = 11.84EE56 pKa = 4.0RR57 pKa = 11.84NSDD60 pKa = 3.16KK61 pKa = 11.23ARR63 pKa = 11.84LVSRR67 pKa = 11.84KK68 pKa = 9.49LLLAAPLFYY77 pKa = 10.69SPPGSEE83 pKa = 3.41TWVRR87 pKa = 11.84TTAVKK92 pKa = 10.06PDD94 pKa = 3.54NASDD98 pKa = 4.45LIEE101 pKa = 5.38DD102 pKa = 3.94VLEE105 pKa = 4.57DD106 pKa = 3.83TLKK109 pKa = 10.18TDD111 pKa = 3.15ATTTPLTTLSQRR123 pKa = 11.84VLKK126 pKa = 10.61ILDD129 pKa = 3.54SSAGDD134 pKa = 3.39SLVSQEE140 pKa = 4.84TILEE144 pKa = 4.08YY145 pKa = 10.99LPIYY149 pKa = 10.3AIIPWRR155 pKa = 11.84ALVKK159 pKa = 10.65SGGVTSSEE167 pKa = 4.14WFSSATGAMTTAIKK181 pKa = 10.54AADD184 pKa = 3.71DD185 pKa = 3.8RR186 pKa = 11.84LRR188 pKa = 11.84QADD191 pKa = 3.88EE192 pKa = 4.1TVKK195 pKa = 11.1NDD197 pKa = 3.03MKK199 pKa = 10.71TWFAQVDD206 pKa = 3.89PTEE209 pKa = 4.58LLTNHH214 pKa = 6.98LAEE217 pKa = 4.94IMACALAHH225 pKa = 5.59SEE227 pKa = 4.11RR228 pKa = 11.84VPPFIAASIQSVWMYY243 pKa = 10.48HH244 pKa = 5.79GLGTYY249 pKa = 10.03IFLRR253 pKa = 11.84EE254 pKa = 3.91AARR257 pKa = 11.84TLGLNSPEE265 pKa = 4.29LLRR268 pKa = 11.84EE269 pKa = 4.33LCCPLLNNQIMALVKK284 pKa = 10.73LMIDD288 pKa = 3.37EE289 pKa = 4.36QANNTPVFPYY299 pKa = 10.35IKK301 pKa = 9.8IINTNFHH308 pKa = 6.49SGVSTKK314 pKa = 10.51RR315 pKa = 11.84CIPLMYY321 pKa = 10.13VLRR324 pKa = 11.84QIICPISPKK333 pKa = 9.36TKK335 pKa = 9.49QGEE338 pKa = 4.71GIWGKK343 pKa = 8.78MCGVPSLDD351 pKa = 3.17VRR353 pKa = 11.84IPLAQAAMRR362 pKa = 11.84LRR364 pKa = 11.84RR365 pKa = 11.84KK366 pKa = 10.03HH367 pKa = 5.74ILTVRR372 pKa = 11.84EE373 pKa = 4.48GEE375 pKa = 4.24TSEE378 pKa = 4.78VMQQILGEE386 pKa = 4.16IRR388 pKa = 11.84DD389 pKa = 4.08TPLGYY394 pKa = 10.4DD395 pKa = 3.51DD396 pKa = 4.46TSSEE400 pKa = 4.1QEE402 pKa = 4.14AVDD405 pKa = 4.78PDD407 pKa = 4.41DD408 pKa = 5.65LGNPPAA414 pKa = 5.56
Molecular weight: 45.91 kDa
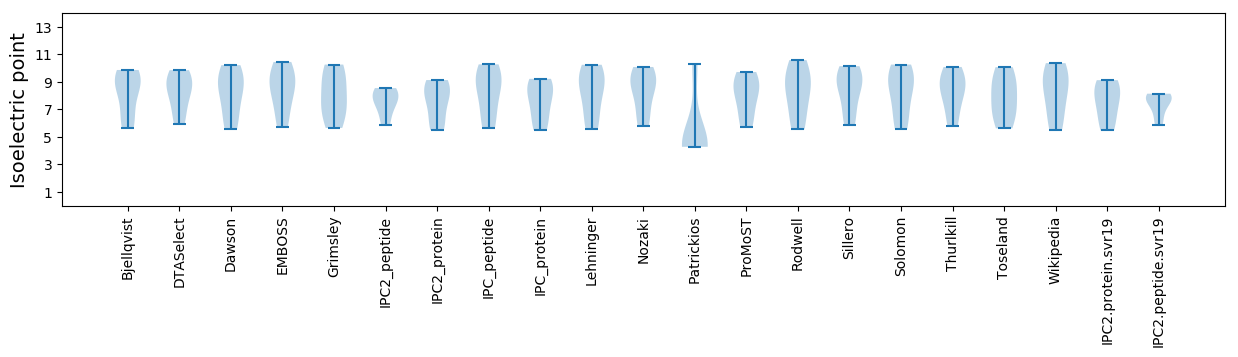
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KMY2|A0A1L3KMY2_9VIRU Uncharacterized protein OS=Beihai barnacle virus 7 OX=1922365 PE=4 SV=1
MM1 pKa = 6.75GQSVSKK7 pKa = 10.28EE8 pKa = 3.48IQEE11 pKa = 4.06RR12 pKa = 11.84WNTQVQALMHH22 pKa = 6.66MIRR25 pKa = 11.84TTIRR29 pKa = 11.84DD30 pKa = 3.26TCDD33 pKa = 2.53ARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84EE38 pKa = 4.18TVEE41 pKa = 3.97TIKK44 pKa = 11.33DD45 pKa = 3.22EE46 pKa = 4.16FRR48 pKa = 11.84RR49 pKa = 11.84SNVHH53 pKa = 6.01IEE55 pKa = 3.9QVYY58 pKa = 10.46KK59 pKa = 10.54ILEE62 pKa = 4.18TAQTVPQPDD71 pKa = 3.78TQLSARR77 pKa = 11.84VEE79 pKa = 4.12NEE81 pKa = 4.29LFNQDD86 pKa = 2.49VWNNAGLGTCEE97 pKa = 4.41PPIKK101 pKa = 9.92IYY103 pKa = 10.91AVLSGVIVLLLALSIMFSVLWRR125 pKa = 11.84IEE127 pKa = 3.89KK128 pKa = 9.9AKK130 pKa = 10.6KK131 pKa = 9.41RR132 pKa = 11.84FRR134 pKa = 11.84RR135 pKa = 11.84TSSFRR140 pKa = 11.84PRR142 pKa = 11.84SILKK146 pKa = 10.18KK147 pKa = 10.14PPSPDD152 pKa = 3.22NVSLSSIRR160 pKa = 11.84TLYY163 pKa = 10.65
MM1 pKa = 6.75GQSVSKK7 pKa = 10.28EE8 pKa = 3.48IQEE11 pKa = 4.06RR12 pKa = 11.84WNTQVQALMHH22 pKa = 6.66MIRR25 pKa = 11.84TTIRR29 pKa = 11.84DD30 pKa = 3.26TCDD33 pKa = 2.53ARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84EE38 pKa = 4.18TVEE41 pKa = 3.97TIKK44 pKa = 11.33DD45 pKa = 3.22EE46 pKa = 4.16FRR48 pKa = 11.84RR49 pKa = 11.84SNVHH53 pKa = 6.01IEE55 pKa = 3.9QVYY58 pKa = 10.46KK59 pKa = 10.54ILEE62 pKa = 4.18TAQTVPQPDD71 pKa = 3.78TQLSARR77 pKa = 11.84VEE79 pKa = 4.12NEE81 pKa = 4.29LFNQDD86 pKa = 2.49VWNNAGLGTCEE97 pKa = 4.41PPIKK101 pKa = 9.92IYY103 pKa = 10.91AVLSGVIVLLLALSIMFSVLWRR125 pKa = 11.84IEE127 pKa = 3.89KK128 pKa = 9.9AKK130 pKa = 10.6KK131 pKa = 9.41RR132 pKa = 11.84FRR134 pKa = 11.84RR135 pKa = 11.84TSSFRR140 pKa = 11.84PRR142 pKa = 11.84SILKK146 pKa = 10.18KK147 pKa = 10.14PPSPDD152 pKa = 3.22NVSLSSIRR160 pKa = 11.84TLYY163 pKa = 10.65
Molecular weight: 18.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3521 |
153 |
1994 |
586.8 |
65.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.993 ± 0.732 | 1.874 ± 0.324 |
4.487 ± 0.524 | 5.283 ± 0.481 |
3.465 ± 0.605 | 4.857 ± 0.463 |
2.215 ± 0.238 | 5.623 ± 0.263 |
4.232 ± 0.319 | 11.843 ± 1.078 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.988 ± 0.152 | 3.55 ± 0.337 |
5.993 ± 0.318 | 3.323 ± 0.292 |
6.816 ± 0.469 | 9.458 ± 0.623 |
7.242 ± 0.622 | 6.93 ± 0.767 |
1.448 ± 0.258 | 3.38 ± 0.347 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
