
Novosphingobium sp. GV010
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Novosphingobium; unclassified Novosphingobium
Average proteome isoelectric point is 6.7
Get precalculated fractions of proteins
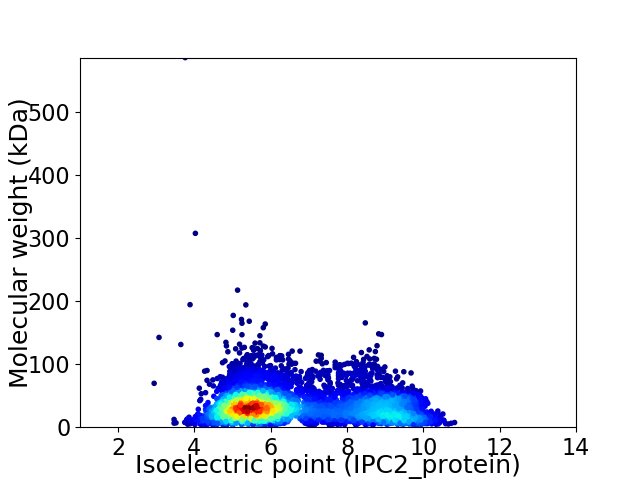
Virtual 2D-PAGE plot for 5048 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2V5C8F3|A0A2V5C8F3_9SPHN DNA-binding transcriptional LysR family regulator OS=Novosphingobium sp. GV010 OX=2135477 GN=C7493_108153 PE=3 SV=1
MM1 pKa = 7.17ATIFRR6 pKa = 11.84SGYY9 pKa = 10.55GDD11 pKa = 3.48WSDD14 pKa = 3.24MADD17 pKa = 3.24WYY19 pKa = 11.15GHH21 pKa = 6.33AVPTAADD28 pKa = 3.41DD29 pKa = 5.14AVFQGNALYY38 pKa = 10.73AGITGVAQAAGLSLADD54 pKa = 3.65GSGIGVGQGGTLTVGDD70 pKa = 4.35GGIALASSAWLQIVGTGTTATTTGTLTLDD99 pKa = 4.03DD100 pKa = 4.46ASVAVMLGEE109 pKa = 4.53EE110 pKa = 5.0GSASDD115 pKa = 3.73VTLSASALDD124 pKa = 3.7SRR126 pKa = 11.84GTIQLHH132 pKa = 5.77AAGGSHH138 pKa = 6.9ARR140 pKa = 11.84LLVSGAAGFGTAGVLTGSVLLDD162 pKa = 3.67GASTIAFGSGQITAIAGDD180 pKa = 3.97LTLSGAQATITSGEE194 pKa = 4.14VAGNSALTGLTSLAGTLTLTGLALTLTGDD223 pKa = 3.96LDD225 pKa = 3.84VDD227 pKa = 3.99GALQLGSSDD236 pKa = 3.51PVASMRR242 pKa = 11.84ATTDD246 pKa = 3.05LTVAGTLTGNGIVQILGTAANGAATLTVGGLAHH279 pKa = 6.67TGFVAITGTANAAAMLVDD297 pKa = 4.23TGAAPATLTGSLLLTDD313 pKa = 4.23HH314 pKa = 7.08ASVHH318 pKa = 5.81LASGTIAQIAQGSTVTLSDD337 pKa = 2.94ATSFVSSEE345 pKa = 4.16ADD347 pKa = 3.18TASNSGLAGLSVVAGALTLNGGGLAITGDD376 pKa = 4.11LGNSGTIRR384 pKa = 11.84LNNGSDD390 pKa = 3.87PDD392 pKa = 3.9TGAATALAATIAGTLTNSGILSLMADD418 pKa = 4.01GSYY421 pKa = 11.41GSGSVGLAVAGIDD434 pKa = 3.5NTPDD438 pKa = 3.35GNIMIAGNADD448 pKa = 3.48TGDD451 pKa = 3.64TAILAVASAAPAALDD466 pKa = 3.74GLWQVGANGSVVFASGGIADD486 pKa = 4.5IVASGQLVLSSASAVFANAADD507 pKa = 3.96TASNSALSSLASIEE521 pKa = 4.03GVLAIASGSVVLGGALNNSGGIYY544 pKa = 10.3LATGSSGPGTAPDD557 pKa = 3.73TASLSIAGVLTNSGTITIGGDD578 pKa = 2.93NSAATLTTGGLINTGSLSVTANVAMAINGLLVNDD612 pKa = 4.31GVMDD616 pKa = 3.96LTLAQRR622 pKa = 11.84LDD624 pKa = 3.78LTGVTALTLGAQASTMLGLAQDD646 pKa = 4.39GPLVSDD652 pKa = 4.06TLAIDD657 pKa = 4.3GGTLTVDD664 pKa = 3.13AGGVPLNAGDD674 pKa = 3.98RR675 pKa = 11.84FTIFSFGAGSANGLFDD691 pKa = 3.82TFSLSSIGAGIGGYY705 pKa = 7.38VQHH708 pKa = 7.19GDD710 pKa = 3.68LLIGLVYY717 pKa = 9.92EE718 pKa = 4.32DD719 pKa = 3.2HH720 pKa = 7.14AGRR723 pKa = 11.84IEE725 pKa = 4.28MVVAEE730 pKa = 5.16APASTADD737 pKa = 3.36SFATSGGAWEE747 pKa = 6.18DD748 pKa = 3.27IAKK751 pKa = 7.08WTQGAPAFYY760 pKa = 9.47AQASIAHH767 pKa = 6.42AAVTLSSDD775 pKa = 3.33ATVSTLALTDD785 pKa = 3.69GATLGVGVLSDD796 pKa = 4.03LSVTGALTVGEE807 pKa = 4.58GTRR810 pKa = 11.84LTVTGGALASAGTQQVDD827 pKa = 2.94GHH829 pKa = 6.89LILTGGAQAFLHH841 pKa = 5.45GTLYY845 pKa = 11.04GGGTIALAGSAVLHH859 pKa = 5.89VEE861 pKa = 4.18GSVAAGLTIDD871 pKa = 4.57FGAAGVGDD879 pKa = 4.41EE880 pKa = 5.28MILTTPGAFDD890 pKa = 3.27GTIAGFAPGDD900 pKa = 3.8TLDD903 pKa = 3.76LAGIAGAHH911 pKa = 5.85LVLGAGTQGGTQLLVEE927 pKa = 5.24DD928 pKa = 4.39GAGTVLVTLAFEE940 pKa = 4.47GAYY943 pKa = 10.04GLSSFTLTQDD953 pKa = 3.21GAGGSLLSLAGNQPSPLTIGAVSNTVVEE981 pKa = 4.63SGAGVAGIATGTALATATDD1000 pKa = 3.6ASFVLTGFTHH1010 pKa = 6.88IGGTVWSEE1018 pKa = 3.38QGTYY1022 pKa = 8.19GTLMLDD1028 pKa = 3.64TATGGLTYY1036 pKa = 10.5TLDD1039 pKa = 3.64NASAATNALSGGAITADD1056 pKa = 3.2RR1057 pKa = 11.84FTIAVTDD1064 pKa = 3.52THH1066 pKa = 6.86GLTTSQAVSFAITGTNDD1083 pKa = 2.94APVVADD1089 pKa = 3.29KK1090 pKa = 9.72TQAVAYY1096 pKa = 9.05NGSAAMTAAALLVGASDD1113 pKa = 4.31AEE1115 pKa = 4.29GDD1117 pKa = 3.67ALSVVGASGAQHH1129 pKa = 6.08GTLSMDD1135 pKa = 3.7ADD1137 pKa = 3.99GTVHH1141 pKa = 6.48YY1142 pKa = 10.33SAFQAYY1148 pKa = 9.19VGADD1152 pKa = 3.16SYY1154 pKa = 11.51QVVVSDD1160 pKa = 3.35SHH1162 pKa = 6.76GATQVEE1168 pKa = 4.87TVKK1171 pKa = 9.58VTVSGATTAAATTTPAYY1188 pKa = 9.55VWAGASTTGQVFDD1201 pKa = 4.02VSGDD1205 pKa = 3.7GLGHH1209 pKa = 7.05RR1210 pKa = 11.84LYY1212 pKa = 10.59ATGGDD1217 pKa = 3.92DD1218 pKa = 4.58TIHH1221 pKa = 6.64GGNGGDD1227 pKa = 4.1ALNGGAGNDD1236 pKa = 3.65IIYY1239 pKa = 10.77GGDD1242 pKa = 3.57GKK1244 pKa = 9.95DD1245 pKa = 3.36TLTGGAGADD1254 pKa = 3.91RR1255 pKa = 11.84VWGGAGADD1263 pKa = 3.18TFVFRR1268 pKa = 11.84PGDD1271 pKa = 3.59MADD1274 pKa = 3.48PALTGGLYY1282 pKa = 10.63DD1283 pKa = 5.02EE1284 pKa = 5.57IMDD1287 pKa = 4.58FSGAGNGYY1295 pKa = 10.06RR1296 pKa = 11.84PGEE1299 pKa = 4.21DD1300 pKa = 2.6MLVFSGFSSAATLTYY1315 pKa = 10.79DD1316 pKa = 4.21VAATQAASDD1325 pKa = 3.96ATAHH1329 pKa = 6.36YY1330 pKa = 8.35YY1331 pKa = 9.66TVTDD1335 pKa = 4.49GAWHH1339 pKa = 6.75GDD1341 pKa = 3.5IIVHH1345 pKa = 4.44YY1346 pKa = 8.81TGNAVLQLGDD1356 pKa = 3.66YY1357 pKa = 11.07GFLL1360 pKa = 3.71
MM1 pKa = 7.17ATIFRR6 pKa = 11.84SGYY9 pKa = 10.55GDD11 pKa = 3.48WSDD14 pKa = 3.24MADD17 pKa = 3.24WYY19 pKa = 11.15GHH21 pKa = 6.33AVPTAADD28 pKa = 3.41DD29 pKa = 5.14AVFQGNALYY38 pKa = 10.73AGITGVAQAAGLSLADD54 pKa = 3.65GSGIGVGQGGTLTVGDD70 pKa = 4.35GGIALASSAWLQIVGTGTTATTTGTLTLDD99 pKa = 4.03DD100 pKa = 4.46ASVAVMLGEE109 pKa = 4.53EE110 pKa = 5.0GSASDD115 pKa = 3.73VTLSASALDD124 pKa = 3.7SRR126 pKa = 11.84GTIQLHH132 pKa = 5.77AAGGSHH138 pKa = 6.9ARR140 pKa = 11.84LLVSGAAGFGTAGVLTGSVLLDD162 pKa = 3.67GASTIAFGSGQITAIAGDD180 pKa = 3.97LTLSGAQATITSGEE194 pKa = 4.14VAGNSALTGLTSLAGTLTLTGLALTLTGDD223 pKa = 3.96LDD225 pKa = 3.84VDD227 pKa = 3.99GALQLGSSDD236 pKa = 3.51PVASMRR242 pKa = 11.84ATTDD246 pKa = 3.05LTVAGTLTGNGIVQILGTAANGAATLTVGGLAHH279 pKa = 6.67TGFVAITGTANAAAMLVDD297 pKa = 4.23TGAAPATLTGSLLLTDD313 pKa = 4.23HH314 pKa = 7.08ASVHH318 pKa = 5.81LASGTIAQIAQGSTVTLSDD337 pKa = 2.94ATSFVSSEE345 pKa = 4.16ADD347 pKa = 3.18TASNSGLAGLSVVAGALTLNGGGLAITGDD376 pKa = 4.11LGNSGTIRR384 pKa = 11.84LNNGSDD390 pKa = 3.87PDD392 pKa = 3.9TGAATALAATIAGTLTNSGILSLMADD418 pKa = 4.01GSYY421 pKa = 11.41GSGSVGLAVAGIDD434 pKa = 3.5NTPDD438 pKa = 3.35GNIMIAGNADD448 pKa = 3.48TGDD451 pKa = 3.64TAILAVASAAPAALDD466 pKa = 3.74GLWQVGANGSVVFASGGIADD486 pKa = 4.5IVASGQLVLSSASAVFANAADD507 pKa = 3.96TASNSALSSLASIEE521 pKa = 4.03GVLAIASGSVVLGGALNNSGGIYY544 pKa = 10.3LATGSSGPGTAPDD557 pKa = 3.73TASLSIAGVLTNSGTITIGGDD578 pKa = 2.93NSAATLTTGGLINTGSLSVTANVAMAINGLLVNDD612 pKa = 4.31GVMDD616 pKa = 3.96LTLAQRR622 pKa = 11.84LDD624 pKa = 3.78LTGVTALTLGAQASTMLGLAQDD646 pKa = 4.39GPLVSDD652 pKa = 4.06TLAIDD657 pKa = 4.3GGTLTVDD664 pKa = 3.13AGGVPLNAGDD674 pKa = 3.98RR675 pKa = 11.84FTIFSFGAGSANGLFDD691 pKa = 3.82TFSLSSIGAGIGGYY705 pKa = 7.38VQHH708 pKa = 7.19GDD710 pKa = 3.68LLIGLVYY717 pKa = 9.92EE718 pKa = 4.32DD719 pKa = 3.2HH720 pKa = 7.14AGRR723 pKa = 11.84IEE725 pKa = 4.28MVVAEE730 pKa = 5.16APASTADD737 pKa = 3.36SFATSGGAWEE747 pKa = 6.18DD748 pKa = 3.27IAKK751 pKa = 7.08WTQGAPAFYY760 pKa = 9.47AQASIAHH767 pKa = 6.42AAVTLSSDD775 pKa = 3.33ATVSTLALTDD785 pKa = 3.69GATLGVGVLSDD796 pKa = 4.03LSVTGALTVGEE807 pKa = 4.58GTRR810 pKa = 11.84LTVTGGALASAGTQQVDD827 pKa = 2.94GHH829 pKa = 6.89LILTGGAQAFLHH841 pKa = 5.45GTLYY845 pKa = 11.04GGGTIALAGSAVLHH859 pKa = 5.89VEE861 pKa = 4.18GSVAAGLTIDD871 pKa = 4.57FGAAGVGDD879 pKa = 4.41EE880 pKa = 5.28MILTTPGAFDD890 pKa = 3.27GTIAGFAPGDD900 pKa = 3.8TLDD903 pKa = 3.76LAGIAGAHH911 pKa = 5.85LVLGAGTQGGTQLLVEE927 pKa = 5.24DD928 pKa = 4.39GAGTVLVTLAFEE940 pKa = 4.47GAYY943 pKa = 10.04GLSSFTLTQDD953 pKa = 3.21GAGGSLLSLAGNQPSPLTIGAVSNTVVEE981 pKa = 4.63SGAGVAGIATGTALATATDD1000 pKa = 3.6ASFVLTGFTHH1010 pKa = 6.88IGGTVWSEE1018 pKa = 3.38QGTYY1022 pKa = 8.19GTLMLDD1028 pKa = 3.64TATGGLTYY1036 pKa = 10.5TLDD1039 pKa = 3.64NASAATNALSGGAITADD1056 pKa = 3.2RR1057 pKa = 11.84FTIAVTDD1064 pKa = 3.52THH1066 pKa = 6.86GLTTSQAVSFAITGTNDD1083 pKa = 2.94APVVADD1089 pKa = 3.29KK1090 pKa = 9.72TQAVAYY1096 pKa = 9.05NGSAAMTAAALLVGASDD1113 pKa = 4.31AEE1115 pKa = 4.29GDD1117 pKa = 3.67ALSVVGASGAQHH1129 pKa = 6.08GTLSMDD1135 pKa = 3.7ADD1137 pKa = 3.99GTVHH1141 pKa = 6.48YY1142 pKa = 10.33SAFQAYY1148 pKa = 9.19VGADD1152 pKa = 3.16SYY1154 pKa = 11.51QVVVSDD1160 pKa = 3.35SHH1162 pKa = 6.76GATQVEE1168 pKa = 4.87TVKK1171 pKa = 9.58VTVSGATTAAATTTPAYY1188 pKa = 9.55VWAGASTTGQVFDD1201 pKa = 4.02VSGDD1205 pKa = 3.7GLGHH1209 pKa = 7.05RR1210 pKa = 11.84LYY1212 pKa = 10.59ATGGDD1217 pKa = 3.92DD1218 pKa = 4.58TIHH1221 pKa = 6.64GGNGGDD1227 pKa = 4.1ALNGGAGNDD1236 pKa = 3.65IIYY1239 pKa = 10.77GGDD1242 pKa = 3.57GKK1244 pKa = 9.95DD1245 pKa = 3.36TLTGGAGADD1254 pKa = 3.91RR1255 pKa = 11.84VWGGAGADD1263 pKa = 3.18TFVFRR1268 pKa = 11.84PGDD1271 pKa = 3.59MADD1274 pKa = 3.48PALTGGLYY1282 pKa = 10.63DD1283 pKa = 5.02EE1284 pKa = 5.57IMDD1287 pKa = 4.58FSGAGNGYY1295 pKa = 10.06RR1296 pKa = 11.84PGEE1299 pKa = 4.21DD1300 pKa = 2.6MLVFSGFSSAATLTYY1315 pKa = 10.79DD1316 pKa = 4.21VAATQAASDD1325 pKa = 3.96ATAHH1329 pKa = 6.36YY1330 pKa = 8.35YY1331 pKa = 9.66TVTDD1335 pKa = 4.49GAWHH1339 pKa = 6.75GDD1341 pKa = 3.5IIVHH1345 pKa = 4.44YY1346 pKa = 8.81TGNAVLQLGDD1356 pKa = 3.66YY1357 pKa = 11.07GFLL1360 pKa = 3.71
Molecular weight: 131.0 kDa
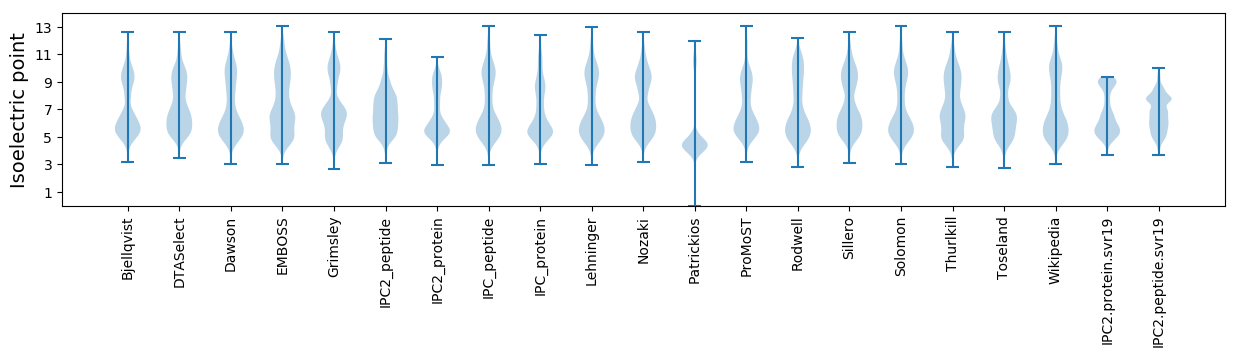
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2V5B5Y2|A0A2V5B5Y2_9SPHN Uncharacterized protein OS=Novosphingobium sp. GV010 OX=2135477 GN=C7493_107132 PE=4 SV=1
MM1 pKa = 8.47AEE3 pKa = 3.94MARR6 pKa = 11.84NGMTSLPAIGFMLATVAMVVLATVMWFVALVTLLAVVGLRR46 pKa = 11.84RR47 pKa = 11.84PLRR50 pKa = 11.84SRR52 pKa = 11.84QGRR55 pKa = 11.84MIRR58 pKa = 11.84MARR61 pKa = 11.84RR62 pKa = 11.84AGSARR67 pKa = 11.84PP68 pKa = 3.43
MM1 pKa = 8.47AEE3 pKa = 3.94MARR6 pKa = 11.84NGMTSLPAIGFMLATVAMVVLATVMWFVALVTLLAVVGLRR46 pKa = 11.84RR47 pKa = 11.84PLRR50 pKa = 11.84SRR52 pKa = 11.84QGRR55 pKa = 11.84MIRR58 pKa = 11.84MARR61 pKa = 11.84RR62 pKa = 11.84AGSARR67 pKa = 11.84PP68 pKa = 3.43
Molecular weight: 7.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1713247 |
26 |
6640 |
339.4 |
36.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.38 ± 0.052 | 0.802 ± 0.01 |
5.78 ± 0.028 | 4.443 ± 0.034 |
3.446 ± 0.021 | 9.266 ± 0.099 |
2.293 ± 0.018 | 4.788 ± 0.022 |
2.624 ± 0.027 | 9.98 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.581 ± 0.018 | 2.614 ± 0.028 |
5.48 ± 0.038 | 3.411 ± 0.021 |
6.928 ± 0.038 | 5.338 ± 0.038 |
5.761 ± 0.038 | 7.301 ± 0.029 |
1.482 ± 0.018 | 2.303 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
