
Streptococcus phage Javan78
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
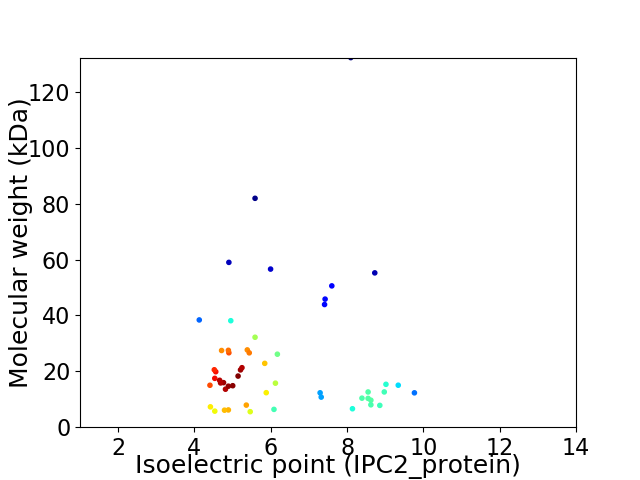
Virtual 2D-PAGE plot for 53 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4D6BEV9|A0A4D6BEV9_9CAUD Minor capsid protein OS=Streptococcus phage Javan78 OX=2548306 GN=Javan78_0011 PE=4 SV=1
MM1 pKa = 7.51NKK3 pKa = 9.68NDD5 pKa = 4.12LFVDD9 pKa = 4.34VASHH13 pKa = 5.34QGYY16 pKa = 10.07NITGILGQMGTTNTIIKK33 pKa = 9.7ISEE36 pKa = 3.99STSYY40 pKa = 11.02INPCLSAQVEE50 pKa = 4.11QSNPIGFYY58 pKa = 9.88HH59 pKa = 5.76YY60 pKa = 10.64AWFGGDD66 pKa = 2.96VAEE69 pKa = 4.77AEE71 pKa = 3.99RR72 pKa = 11.84EE73 pKa = 3.78ARR75 pKa = 11.84YY76 pKa = 9.49FIDD79 pKa = 4.55NVPQKK84 pKa = 11.01VKK86 pKa = 10.38YY87 pKa = 10.42LCLDD91 pKa = 3.96YY92 pKa = 10.92EE93 pKa = 4.44DD94 pKa = 5.0HH95 pKa = 7.98ASGDD99 pKa = 3.59KK100 pKa = 10.11QANTDD105 pKa = 2.83ACLRR109 pKa = 11.84FMEE112 pKa = 4.24ILKK115 pKa = 10.54EE116 pKa = 3.95NGYY119 pKa = 10.25EE120 pKa = 4.01PIYY123 pKa = 10.7YY124 pKa = 9.76SYY126 pKa = 11.64KK127 pKa = 10.54PFTLNNIYY135 pKa = 10.45YY136 pKa = 8.43EE137 pKa = 4.23QILARR142 pKa = 11.84FPNSLWIAGYY152 pKa = 10.38GLNDD156 pKa = 3.33GNADD160 pKa = 3.61FNYY163 pKa = 8.87FPSMDD168 pKa = 4.52GIRR171 pKa = 11.84WWQYY175 pKa = 10.03SSNPFDD181 pKa = 4.42KK182 pKa = 11.23NIVLLDD188 pKa = 3.94DD189 pKa = 4.18EE190 pKa = 5.68EE191 pKa = 4.64EE192 pKa = 4.62DD193 pKa = 4.11NVISKK198 pKa = 10.66DD199 pKa = 3.49AVKK202 pKa = 10.71SLNTVADD209 pKa = 3.93EE210 pKa = 4.71VIQGLWGNGQEE221 pKa = 5.08RR222 pKa = 11.84FDD224 pKa = 3.84NLTNAGYY231 pKa = 9.79NAQAVQDD238 pKa = 4.13KK239 pKa = 11.44VNDD242 pKa = 3.94LLNAEE247 pKa = 4.45NTSKK251 pKa = 11.19DD252 pKa = 3.75LDD254 pKa = 3.81TLANEE259 pKa = 4.39VLQGLWGNGQEE270 pKa = 5.14RR271 pKa = 11.84FDD273 pKa = 3.71KK274 pKa = 11.2LEE276 pKa = 3.98NAGYY280 pKa = 9.79DD281 pKa = 3.43AQAVQDD287 pKa = 4.1RR288 pKa = 11.84VNSLLSGGNTADD300 pKa = 4.36LDD302 pKa = 4.12TVANEE307 pKa = 4.18VVQGLWGNGQEE318 pKa = 5.08RR319 pKa = 11.84FDD321 pKa = 3.84NLTNAGYY328 pKa = 9.89DD329 pKa = 3.29AQAVQNRR336 pKa = 11.84VNEE339 pKa = 4.16LLSS342 pKa = 3.58
MM1 pKa = 7.51NKK3 pKa = 9.68NDD5 pKa = 4.12LFVDD9 pKa = 4.34VASHH13 pKa = 5.34QGYY16 pKa = 10.07NITGILGQMGTTNTIIKK33 pKa = 9.7ISEE36 pKa = 3.99STSYY40 pKa = 11.02INPCLSAQVEE50 pKa = 4.11QSNPIGFYY58 pKa = 9.88HH59 pKa = 5.76YY60 pKa = 10.64AWFGGDD66 pKa = 2.96VAEE69 pKa = 4.77AEE71 pKa = 3.99RR72 pKa = 11.84EE73 pKa = 3.78ARR75 pKa = 11.84YY76 pKa = 9.49FIDD79 pKa = 4.55NVPQKK84 pKa = 11.01VKK86 pKa = 10.38YY87 pKa = 10.42LCLDD91 pKa = 3.96YY92 pKa = 10.92EE93 pKa = 4.44DD94 pKa = 5.0HH95 pKa = 7.98ASGDD99 pKa = 3.59KK100 pKa = 10.11QANTDD105 pKa = 2.83ACLRR109 pKa = 11.84FMEE112 pKa = 4.24ILKK115 pKa = 10.54EE116 pKa = 3.95NGYY119 pKa = 10.25EE120 pKa = 4.01PIYY123 pKa = 10.7YY124 pKa = 9.76SYY126 pKa = 11.64KK127 pKa = 10.54PFTLNNIYY135 pKa = 10.45YY136 pKa = 8.43EE137 pKa = 4.23QILARR142 pKa = 11.84FPNSLWIAGYY152 pKa = 10.38GLNDD156 pKa = 3.33GNADD160 pKa = 3.61FNYY163 pKa = 8.87FPSMDD168 pKa = 4.52GIRR171 pKa = 11.84WWQYY175 pKa = 10.03SSNPFDD181 pKa = 4.42KK182 pKa = 11.23NIVLLDD188 pKa = 3.94DD189 pKa = 4.18EE190 pKa = 5.68EE191 pKa = 4.64EE192 pKa = 4.62DD193 pKa = 4.11NVISKK198 pKa = 10.66DD199 pKa = 3.49AVKK202 pKa = 10.71SLNTVADD209 pKa = 3.93EE210 pKa = 4.71VIQGLWGNGQEE221 pKa = 5.08RR222 pKa = 11.84FDD224 pKa = 3.84NLTNAGYY231 pKa = 9.79NAQAVQDD238 pKa = 4.13KK239 pKa = 11.44VNDD242 pKa = 3.94LLNAEE247 pKa = 4.45NTSKK251 pKa = 11.19DD252 pKa = 3.75LDD254 pKa = 3.81TLANEE259 pKa = 4.39VLQGLWGNGQEE270 pKa = 5.14RR271 pKa = 11.84FDD273 pKa = 3.71KK274 pKa = 11.2LEE276 pKa = 3.98NAGYY280 pKa = 9.79DD281 pKa = 3.43AQAVQDD287 pKa = 4.1RR288 pKa = 11.84VNSLLSGGNTADD300 pKa = 4.36LDD302 pKa = 4.12TVANEE307 pKa = 4.18VVQGLWGNGQEE318 pKa = 5.08RR319 pKa = 11.84FDD321 pKa = 3.84NLTNAGYY328 pKa = 9.89DD329 pKa = 3.29AQAVQNRR336 pKa = 11.84VNEE339 pKa = 4.16LLSS342 pKa = 3.58
Molecular weight: 38.39 kDa
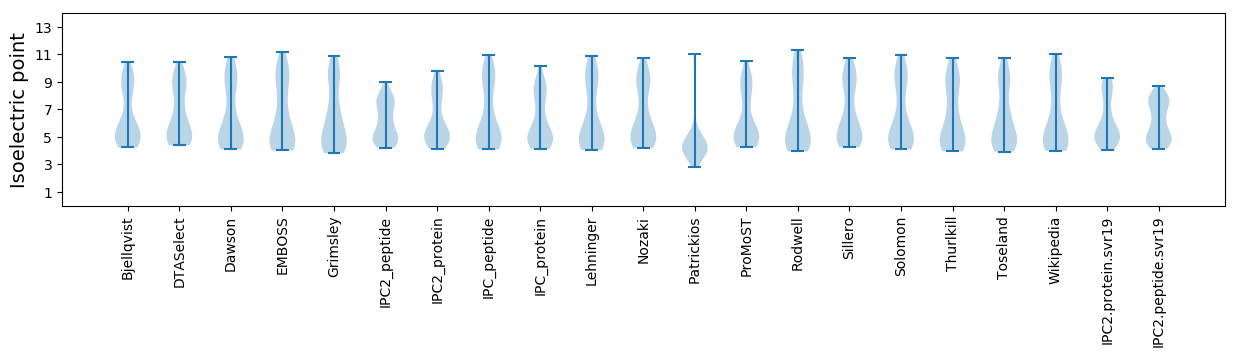
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D6BEH9|A0A4D6BEH9_9CAUD Terminase small subunit OS=Streptococcus phage Javan78 OX=2548306 GN=Javan78_0022 PE=4 SV=1
MM1 pKa = 7.72ADD3 pKa = 3.16VRR5 pKa = 11.84VSIDD9 pKa = 3.26LAGVEE14 pKa = 4.86KK15 pKa = 10.52KK16 pKa = 10.72VSPQAMQRR24 pKa = 11.84GKK26 pKa = 9.98IAAGSEE32 pKa = 3.87ALLIMDD38 pKa = 4.07SSVPLRR44 pKa = 11.84AGGGALRR51 pKa = 11.84ASGRR55 pKa = 11.84VEE57 pKa = 3.73PNGNVSYY64 pKa = 9.14NTVYY68 pKa = 10.96ARR70 pKa = 11.84AQFHH74 pKa = 5.45GTNGIVVFRR83 pKa = 11.84KK84 pKa = 8.49YY85 pKa = 6.16TTSGTGSRR93 pKa = 11.84WDD95 pKa = 3.86KK96 pKa = 10.38PLKK99 pKa = 10.93ANIDD103 pKa = 3.85KK104 pKa = 10.78LKK106 pKa = 10.34KK107 pKa = 10.11AAIKK111 pKa = 10.82GMGIRR116 pKa = 4.21
MM1 pKa = 7.72ADD3 pKa = 3.16VRR5 pKa = 11.84VSIDD9 pKa = 3.26LAGVEE14 pKa = 4.86KK15 pKa = 10.52KK16 pKa = 10.72VSPQAMQRR24 pKa = 11.84GKK26 pKa = 9.98IAAGSEE32 pKa = 3.87ALLIMDD38 pKa = 4.07SSVPLRR44 pKa = 11.84AGGGALRR51 pKa = 11.84ASGRR55 pKa = 11.84VEE57 pKa = 3.73PNGNVSYY64 pKa = 9.14NTVYY68 pKa = 10.96ARR70 pKa = 11.84AQFHH74 pKa = 5.45GTNGIVVFRR83 pKa = 11.84KK84 pKa = 8.49YY85 pKa = 6.16TTSGTGSRR93 pKa = 11.84WDD95 pKa = 3.86KK96 pKa = 10.38PLKK99 pKa = 10.93ANIDD103 pKa = 3.85KK104 pKa = 10.78LKK106 pKa = 10.34KK107 pKa = 10.11AAIKK111 pKa = 10.82GMGIRR116 pKa = 4.21
Molecular weight: 12.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
11218 |
48 |
1293 |
211.7 |
23.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.247 ± 1.036 | 0.526 ± 0.106 |
6.276 ± 0.423 | 6.864 ± 0.537 |
4.172 ± 0.218 | 6.605 ± 0.585 |
1.373 ± 0.14 | 7.31 ± 0.232 |
8.121 ± 0.457 | 8.388 ± 0.27 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.469 ± 0.261 | 6.106 ± 0.345 |
2.549 ± 0.182 | 4.511 ± 0.236 |
3.601 ± 0.382 | 6.4 ± 0.465 |
5.999 ± 0.237 | 6.463 ± 0.167 |
1.007 ± 0.133 | 4.011 ± 0.414 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
