
Alphaproteobacteria bacterium PA3
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; unclassified Alphaproteobacteria
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
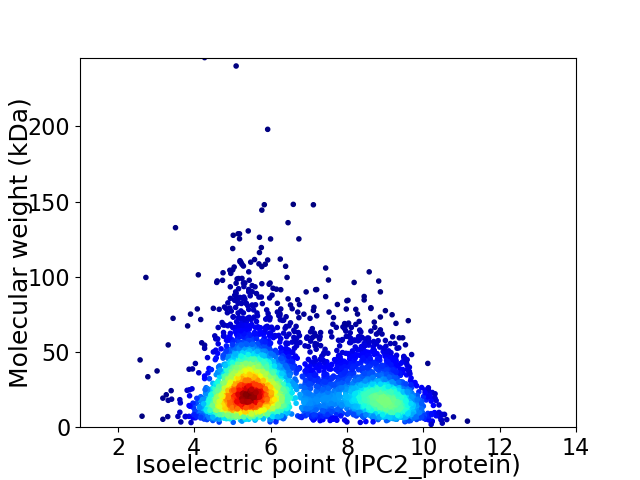
Virtual 2D-PAGE plot for 4749 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A257JLY2|A0A257JLY2_9PROT Uncharacterized protein OS=Alphaproteobacteria bacterium PA3 OX=2015571 GN=CFE32_14480 PE=3 SV=1
MM1 pKa = 7.76CIVCQAKK8 pKa = 10.47SILKK12 pKa = 9.6PDD14 pKa = 3.42YY15 pKa = 10.56EE16 pKa = 5.12LIFGSGMNDD25 pKa = 2.95VPAAAALASKK35 pKa = 10.47EE36 pKa = 3.9PDD38 pKa = 3.46YY39 pKa = 11.71LEE41 pKa = 4.29TANATTSNTATSDD54 pKa = 3.44QNINGVLANTRR65 pKa = 11.84FSSLTLTFAFPQLATDD81 pKa = 3.74YY82 pKa = 8.49EE83 pKa = 4.35ANYY86 pKa = 10.5ASVSATTSQVIGSFSPITAAMQTAARR112 pKa = 11.84FTFEE116 pKa = 5.18LASQLTGLTITEE128 pKa = 4.46GPAATANFRR137 pKa = 11.84LGLFEE142 pKa = 4.56QNADD146 pKa = 3.19GTGPRR151 pKa = 11.84TAFAYY156 pKa = 10.48LPTSSIVGGDD166 pKa = 2.87SWYY169 pKa = 11.16SNTSSFTSPTRR180 pKa = 11.84GNYY183 pKa = 8.41AWATILHH190 pKa = 6.6EE191 pKa = 4.78IGHH194 pKa = 6.09NLGLKK199 pKa = 9.7HH200 pKa = 5.41GHH202 pKa = 5.29EE203 pKa = 4.68TGGPAGVAMAANRR216 pKa = 11.84NSMEE220 pKa = 4.24FSLMTYY226 pKa = 10.71NSFIGDD232 pKa = 3.49DD233 pKa = 3.44TAGYY237 pKa = 8.49TNEE240 pKa = 3.78RR241 pKa = 11.84WGFAQTFMMYY251 pKa = 10.36DD252 pKa = 3.5IAALQHH258 pKa = 6.13MYY260 pKa = 10.81GADD263 pKa = 3.03FTMNNTNSIYY273 pKa = 10.37TFSEE277 pKa = 4.15TTGEE281 pKa = 3.98MFINGVGQGAPGGNRR296 pKa = 11.84VFLTIWDD303 pKa = 3.91GGGTDD308 pKa = 3.99TYY310 pKa = 11.6DD311 pKa = 3.3FSNYY315 pKa = 7.17TTTQLIDD322 pKa = 3.69LRR324 pKa = 11.84PSFWSKK330 pKa = 10.65FSDD333 pKa = 3.55VQTANLGSGNFSRR346 pKa = 11.84GNVFNALQYY355 pKa = 10.89QNDD358 pKa = 3.54GRR360 pKa = 11.84SLIEE364 pKa = 3.76NAFGGSGNDD373 pKa = 4.51DD374 pKa = 2.84IRR376 pKa = 11.84GNQAANTLEE385 pKa = 4.13GRR387 pKa = 11.84DD388 pKa = 3.99GNDD391 pKa = 4.14LINGGSGADD400 pKa = 3.48FLYY403 pKa = 10.71GQVGNDD409 pKa = 3.42TLIGDD414 pKa = 4.25FVVTNSVAANPGVGFTGLAIQTLAQGSAPTSQSTALNLTQNFGLFSNSDD463 pKa = 3.19IGDD466 pKa = 3.93STTSPHH472 pKa = 4.75TTISASGNGAAHH484 pKa = 7.79WFRR487 pKa = 11.84VDD489 pKa = 2.89IASADD494 pKa = 3.53VTITLDD500 pKa = 3.15IDD502 pKa = 3.59ATTGGLDD509 pKa = 3.18SFVRR513 pKa = 11.84LFFLAPGGTIATKK526 pKa = 10.47VAEE529 pKa = 4.46NDD531 pKa = 4.02DD532 pKa = 4.2GSTSAGGGGSATNTDD547 pKa = 3.12SYY549 pKa = 11.07LTYY552 pKa = 10.04LTLQPGTYY560 pKa = 8.78FAVVGTYY567 pKa = 9.78QDD569 pKa = 2.94IDD571 pKa = 3.59ALASGTGYY579 pKa = 8.47TLHH582 pKa = 7.04ISVRR586 pKa = 11.84GEE588 pKa = 3.78VSTTGLGDD596 pKa = 3.52GGTAGDD602 pKa = 3.7DD603 pKa = 3.29QLYY606 pKa = 10.7GGEE609 pKa = 4.62GNDD612 pKa = 3.73TLIGGLGADD621 pKa = 3.74RR622 pKa = 11.84LEE624 pKa = 4.56GQNGVDD630 pKa = 3.18TASYY634 pKa = 10.74ANAATGVYY642 pKa = 10.22VGFGDD647 pKa = 3.64SGNATGEE654 pKa = 4.12AFGDD658 pKa = 3.64TFFSIEE664 pKa = 4.28NLTGSAFSDD673 pKa = 3.06ILSWDD678 pKa = 3.19NNANVISGLAGQDD691 pKa = 3.22QLFGNGGNDD700 pKa = 3.23TLLGGTEE707 pKa = 4.54DD708 pKa = 4.42DD709 pKa = 4.42VLTGGQGGDD718 pKa = 3.62VLNGEE723 pKa = 4.75AGFDD727 pKa = 3.62TASYY731 pKa = 9.54AASAAAIYY739 pKa = 10.38LFAGDD744 pKa = 3.73NGNWTGDD751 pKa = 3.32AFGDD755 pKa = 3.64TFISIEE761 pKa = 4.2RR762 pKa = 11.84VDD764 pKa = 3.64GSNFHH769 pKa = 7.04DD770 pKa = 4.94LIGFSEE776 pKa = 4.63FNDD779 pKa = 3.62EE780 pKa = 4.37VRR782 pKa = 11.84GLAGNDD788 pKa = 3.16QLYY791 pKa = 11.02GLGGDD796 pKa = 4.02DD797 pKa = 3.79VLLGGEE803 pKa = 4.91GRR805 pKa = 11.84DD806 pKa = 3.8LLNGGSGTDD815 pKa = 3.26TASYY819 pKa = 8.22ATSAAAIYY827 pKa = 10.07LVAGDD832 pKa = 3.93EE833 pKa = 4.61GNWTGDD839 pKa = 3.16AFGDD843 pKa = 3.67TLISIEE849 pKa = 4.58RR850 pKa = 11.84VDD852 pKa = 3.7GSNFNDD858 pKa = 3.96IIGLSDD864 pKa = 3.52LADD867 pKa = 3.66EE868 pKa = 4.69VRR870 pKa = 11.84GLSGNDD876 pKa = 3.07QLFGLGGNDD885 pKa = 3.15ILIGGIGRR893 pKa = 11.84DD894 pKa = 3.64VLNGGTGTDD903 pKa = 3.21TASYY907 pKa = 8.21STAAAGVYY915 pKa = 10.22AFAGDD920 pKa = 3.39WGNFTGDD927 pKa = 3.26AAGDD931 pKa = 3.7TFTSIEE937 pKa = 4.33NLTGSAFNDD946 pKa = 3.25ILGMDD951 pKa = 4.56GGNNVIDD958 pKa = 4.47GGAGDD963 pKa = 5.38DD964 pKa = 3.69VLFGQGGGDD973 pKa = 3.68TLIGGAGFDD982 pKa = 3.55TASYY986 pKa = 8.61ATATSGVYY994 pKa = 9.67VFYY997 pKa = 10.98GDD999 pKa = 3.18WGSFTGDD1006 pKa = 2.99ARR1008 pKa = 11.84GDD1010 pKa = 3.48TMFSVEE1016 pKa = 5.27SILGSNFNDD1025 pKa = 4.0IIGMDD1030 pKa = 3.91DD1031 pKa = 4.03FANTLAGGGGDD1042 pKa = 4.25DD1043 pKa = 3.37ILYY1046 pKa = 10.95GEE1048 pKa = 4.87GGGDD1052 pKa = 3.15RR1053 pKa = 11.84LFGNAGFDD1061 pKa = 3.53TVSYY1065 pKa = 10.79AFAPSGVYY1073 pKa = 10.03ILFSDD1078 pKa = 4.7LGNATGDD1085 pKa = 3.41AAGDD1089 pKa = 4.04SEE1091 pKa = 4.8FSSIEE1096 pKa = 4.29CIVGSAFDD1104 pKa = 5.5DD1105 pKa = 4.35IIGMGDD1111 pKa = 3.37FNDD1114 pKa = 3.97TLGGGGGNDD1123 pKa = 2.82VLMGNGGGDD1132 pKa = 2.78RR1133 pKa = 11.84FFGNEE1138 pKa = 3.49GFDD1141 pKa = 3.52TVSYY1145 pKa = 10.3GLAGSGVYY1153 pKa = 10.06IFFADD1158 pKa = 4.31LGNATGDD1165 pKa = 3.35ARR1167 pKa = 11.84GDD1169 pKa = 3.84SEE1171 pKa = 4.25FSSIEE1176 pKa = 4.24SIVGSNFNDD1185 pKa = 3.81VIGMGDD1191 pKa = 3.45FADD1194 pKa = 4.55TIIGGGGNDD1203 pKa = 3.4ILLGMGGNDD1212 pKa = 4.06TLWGGAGNDD1221 pKa = 3.34LFAYY1225 pKa = 9.43TSFFFGTDD1233 pKa = 3.7TIRR1236 pKa = 11.84DD1237 pKa = 3.89FQDD1240 pKa = 3.32GLDD1243 pKa = 4.16RR1244 pKa = 11.84IDD1246 pKa = 4.07FSRR1249 pKa = 11.84LAGATYY1255 pKa = 10.49SGLAITSVAGGTQVALGTSTILLSGINTSQITAADD1290 pKa = 3.71FLFAA1294 pKa = 5.89
MM1 pKa = 7.76CIVCQAKK8 pKa = 10.47SILKK12 pKa = 9.6PDD14 pKa = 3.42YY15 pKa = 10.56EE16 pKa = 5.12LIFGSGMNDD25 pKa = 2.95VPAAAALASKK35 pKa = 10.47EE36 pKa = 3.9PDD38 pKa = 3.46YY39 pKa = 11.71LEE41 pKa = 4.29TANATTSNTATSDD54 pKa = 3.44QNINGVLANTRR65 pKa = 11.84FSSLTLTFAFPQLATDD81 pKa = 3.74YY82 pKa = 8.49EE83 pKa = 4.35ANYY86 pKa = 10.5ASVSATTSQVIGSFSPITAAMQTAARR112 pKa = 11.84FTFEE116 pKa = 5.18LASQLTGLTITEE128 pKa = 4.46GPAATANFRR137 pKa = 11.84LGLFEE142 pKa = 4.56QNADD146 pKa = 3.19GTGPRR151 pKa = 11.84TAFAYY156 pKa = 10.48LPTSSIVGGDD166 pKa = 2.87SWYY169 pKa = 11.16SNTSSFTSPTRR180 pKa = 11.84GNYY183 pKa = 8.41AWATILHH190 pKa = 6.6EE191 pKa = 4.78IGHH194 pKa = 6.09NLGLKK199 pKa = 9.7HH200 pKa = 5.41GHH202 pKa = 5.29EE203 pKa = 4.68TGGPAGVAMAANRR216 pKa = 11.84NSMEE220 pKa = 4.24FSLMTYY226 pKa = 10.71NSFIGDD232 pKa = 3.49DD233 pKa = 3.44TAGYY237 pKa = 8.49TNEE240 pKa = 3.78RR241 pKa = 11.84WGFAQTFMMYY251 pKa = 10.36DD252 pKa = 3.5IAALQHH258 pKa = 6.13MYY260 pKa = 10.81GADD263 pKa = 3.03FTMNNTNSIYY273 pKa = 10.37TFSEE277 pKa = 4.15TTGEE281 pKa = 3.98MFINGVGQGAPGGNRR296 pKa = 11.84VFLTIWDD303 pKa = 3.91GGGTDD308 pKa = 3.99TYY310 pKa = 11.6DD311 pKa = 3.3FSNYY315 pKa = 7.17TTTQLIDD322 pKa = 3.69LRR324 pKa = 11.84PSFWSKK330 pKa = 10.65FSDD333 pKa = 3.55VQTANLGSGNFSRR346 pKa = 11.84GNVFNALQYY355 pKa = 10.89QNDD358 pKa = 3.54GRR360 pKa = 11.84SLIEE364 pKa = 3.76NAFGGSGNDD373 pKa = 4.51DD374 pKa = 2.84IRR376 pKa = 11.84GNQAANTLEE385 pKa = 4.13GRR387 pKa = 11.84DD388 pKa = 3.99GNDD391 pKa = 4.14LINGGSGADD400 pKa = 3.48FLYY403 pKa = 10.71GQVGNDD409 pKa = 3.42TLIGDD414 pKa = 4.25FVVTNSVAANPGVGFTGLAIQTLAQGSAPTSQSTALNLTQNFGLFSNSDD463 pKa = 3.19IGDD466 pKa = 3.93STTSPHH472 pKa = 4.75TTISASGNGAAHH484 pKa = 7.79WFRR487 pKa = 11.84VDD489 pKa = 2.89IASADD494 pKa = 3.53VTITLDD500 pKa = 3.15IDD502 pKa = 3.59ATTGGLDD509 pKa = 3.18SFVRR513 pKa = 11.84LFFLAPGGTIATKK526 pKa = 10.47VAEE529 pKa = 4.46NDD531 pKa = 4.02DD532 pKa = 4.2GSTSAGGGGSATNTDD547 pKa = 3.12SYY549 pKa = 11.07LTYY552 pKa = 10.04LTLQPGTYY560 pKa = 8.78FAVVGTYY567 pKa = 9.78QDD569 pKa = 2.94IDD571 pKa = 3.59ALASGTGYY579 pKa = 8.47TLHH582 pKa = 7.04ISVRR586 pKa = 11.84GEE588 pKa = 3.78VSTTGLGDD596 pKa = 3.52GGTAGDD602 pKa = 3.7DD603 pKa = 3.29QLYY606 pKa = 10.7GGEE609 pKa = 4.62GNDD612 pKa = 3.73TLIGGLGADD621 pKa = 3.74RR622 pKa = 11.84LEE624 pKa = 4.56GQNGVDD630 pKa = 3.18TASYY634 pKa = 10.74ANAATGVYY642 pKa = 10.22VGFGDD647 pKa = 3.64SGNATGEE654 pKa = 4.12AFGDD658 pKa = 3.64TFFSIEE664 pKa = 4.28NLTGSAFSDD673 pKa = 3.06ILSWDD678 pKa = 3.19NNANVISGLAGQDD691 pKa = 3.22QLFGNGGNDD700 pKa = 3.23TLLGGTEE707 pKa = 4.54DD708 pKa = 4.42DD709 pKa = 4.42VLTGGQGGDD718 pKa = 3.62VLNGEE723 pKa = 4.75AGFDD727 pKa = 3.62TASYY731 pKa = 9.54AASAAAIYY739 pKa = 10.38LFAGDD744 pKa = 3.73NGNWTGDD751 pKa = 3.32AFGDD755 pKa = 3.64TFISIEE761 pKa = 4.2RR762 pKa = 11.84VDD764 pKa = 3.64GSNFHH769 pKa = 7.04DD770 pKa = 4.94LIGFSEE776 pKa = 4.63FNDD779 pKa = 3.62EE780 pKa = 4.37VRR782 pKa = 11.84GLAGNDD788 pKa = 3.16QLYY791 pKa = 11.02GLGGDD796 pKa = 4.02DD797 pKa = 3.79VLLGGEE803 pKa = 4.91GRR805 pKa = 11.84DD806 pKa = 3.8LLNGGSGTDD815 pKa = 3.26TASYY819 pKa = 8.22ATSAAAIYY827 pKa = 10.07LVAGDD832 pKa = 3.93EE833 pKa = 4.61GNWTGDD839 pKa = 3.16AFGDD843 pKa = 3.67TLISIEE849 pKa = 4.58RR850 pKa = 11.84VDD852 pKa = 3.7GSNFNDD858 pKa = 3.96IIGLSDD864 pKa = 3.52LADD867 pKa = 3.66EE868 pKa = 4.69VRR870 pKa = 11.84GLSGNDD876 pKa = 3.07QLFGLGGNDD885 pKa = 3.15ILIGGIGRR893 pKa = 11.84DD894 pKa = 3.64VLNGGTGTDD903 pKa = 3.21TASYY907 pKa = 8.21STAAAGVYY915 pKa = 10.22AFAGDD920 pKa = 3.39WGNFTGDD927 pKa = 3.26AAGDD931 pKa = 3.7TFTSIEE937 pKa = 4.33NLTGSAFNDD946 pKa = 3.25ILGMDD951 pKa = 4.56GGNNVIDD958 pKa = 4.47GGAGDD963 pKa = 5.38DD964 pKa = 3.69VLFGQGGGDD973 pKa = 3.68TLIGGAGFDD982 pKa = 3.55TASYY986 pKa = 8.61ATATSGVYY994 pKa = 9.67VFYY997 pKa = 10.98GDD999 pKa = 3.18WGSFTGDD1006 pKa = 2.99ARR1008 pKa = 11.84GDD1010 pKa = 3.48TMFSVEE1016 pKa = 5.27SILGSNFNDD1025 pKa = 4.0IIGMDD1030 pKa = 3.91DD1031 pKa = 4.03FANTLAGGGGDD1042 pKa = 4.25DD1043 pKa = 3.37ILYY1046 pKa = 10.95GEE1048 pKa = 4.87GGGDD1052 pKa = 3.15RR1053 pKa = 11.84LFGNAGFDD1061 pKa = 3.53TVSYY1065 pKa = 10.79AFAPSGVYY1073 pKa = 10.03ILFSDD1078 pKa = 4.7LGNATGDD1085 pKa = 3.41AAGDD1089 pKa = 4.04SEE1091 pKa = 4.8FSSIEE1096 pKa = 4.29CIVGSAFDD1104 pKa = 5.5DD1105 pKa = 4.35IIGMGDD1111 pKa = 3.37FNDD1114 pKa = 3.97TLGGGGGNDD1123 pKa = 2.82VLMGNGGGDD1132 pKa = 2.78RR1133 pKa = 11.84FFGNEE1138 pKa = 3.49GFDD1141 pKa = 3.52TVSYY1145 pKa = 10.3GLAGSGVYY1153 pKa = 10.06IFFADD1158 pKa = 4.31LGNATGDD1165 pKa = 3.35ARR1167 pKa = 11.84GDD1169 pKa = 3.84SEE1171 pKa = 4.25FSSIEE1176 pKa = 4.24SIVGSNFNDD1185 pKa = 3.81VIGMGDD1191 pKa = 3.45FADD1194 pKa = 4.55TIIGGGGNDD1203 pKa = 3.4ILLGMGGNDD1212 pKa = 4.06TLWGGAGNDD1221 pKa = 3.34LFAYY1225 pKa = 9.43TSFFFGTDD1233 pKa = 3.7TIRR1236 pKa = 11.84DD1237 pKa = 3.89FQDD1240 pKa = 3.32GLDD1243 pKa = 4.16RR1244 pKa = 11.84IDD1246 pKa = 4.07FSRR1249 pKa = 11.84LAGATYY1255 pKa = 10.49SGLAITSVAGGTQVALGTSTILLSGINTSQITAADD1290 pKa = 3.71FLFAA1294 pKa = 5.89
Molecular weight: 132.66 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A257JVV1|A0A257JVV1_9PROT Heat-inducible transcription repressor HrcA OS=Alphaproteobacteria bacterium PA3 OX=2015571 GN=hrcA PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84ARR12 pKa = 11.84MATKK16 pKa = 10.32GGRR19 pKa = 11.84LVLARR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84AKK28 pKa = 9.93GRR30 pKa = 11.84KK31 pKa = 8.85RR32 pKa = 11.84LSAA35 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84ARR12 pKa = 11.84MATKK16 pKa = 10.32GGRR19 pKa = 11.84LVLARR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84AKK28 pKa = 9.93GRR30 pKa = 11.84KK31 pKa = 8.85RR32 pKa = 11.84LSAA35 pKa = 3.96
Molecular weight: 4.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1247484 |
16 |
2327 |
262.7 |
28.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.68 ± 0.054 | 0.853 ± 0.013 |
5.637 ± 0.032 | 5.318 ± 0.038 |
3.789 ± 0.023 | 8.573 ± 0.046 |
1.879 ± 0.017 | 5.069 ± 0.026 |
3.556 ± 0.027 | 10.071 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.372 ± 0.021 | 2.851 ± 0.026 |
5.246 ± 0.026 | 3.568 ± 0.025 |
6.524 ± 0.033 | 5.805 ± 0.027 |
5.536 ± 0.033 | 7.079 ± 0.029 |
1.445 ± 0.017 | 2.146 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
