
Gammapapillomavirus 13
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus
Average proteome isoelectric point is 5.95
Get precalculated fractions of proteins
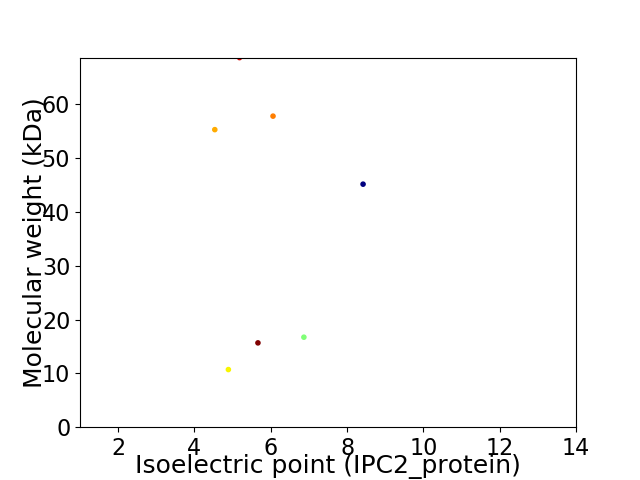
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2D2ALR9|A0A2D2ALR9_9PAPI Protein E6 OS=Gammapapillomavirus 13 OX=1513258 GN=E6 PE=3 SV=1
MM1 pKa = 7.24SRR3 pKa = 11.84ARR5 pKa = 11.84RR6 pKa = 11.84TKK8 pKa = 9.73RR9 pKa = 11.84ASVQDD14 pKa = 4.7LYY16 pKa = 10.98TSCKK20 pKa = 10.4LGGDD24 pKa = 3.74CPEE27 pKa = 4.09DD28 pKa = 3.27VKK30 pKa = 11.65NKK32 pKa = 10.07VEE34 pKa = 4.07NTTLADD40 pKa = 3.79RR41 pKa = 11.84LLQILGSVVYY51 pKa = 9.79FGQLGIGSGKK61 pKa = 8.28GTGGITGYY69 pKa = 10.59RR70 pKa = 11.84PLQNTPRR77 pKa = 11.84VGTNRR82 pKa = 11.84PIEE85 pKa = 4.43SIRR88 pKa = 11.84PNIPIDD94 pKa = 3.64PLGGSDD100 pKa = 4.51IIPPTVIDD108 pKa = 3.41ATAPSVVPLTEE119 pKa = 3.96TGTPEE124 pKa = 3.68VNIISSGTGTNIEE137 pKa = 4.59LGEE140 pKa = 4.23LDD142 pKa = 4.39IITNVDD148 pKa = 3.44PTSTAVGTAEE158 pKa = 4.25HH159 pKa = 6.95PAVIDD164 pKa = 3.6VTDD167 pKa = 4.05EE168 pKa = 4.07GTSVIDD174 pKa = 3.94VQSGPPPPKK183 pKa = 9.87RR184 pKa = 11.84LQIDD188 pKa = 3.75SSLKK192 pKa = 8.95PYY194 pKa = 10.22EE195 pKa = 4.68HH196 pKa = 7.43IEE198 pKa = 4.03LTVYY202 pKa = 10.36PPYY205 pKa = 10.47NQSDD209 pKa = 3.86PNVNVFVDD217 pKa = 3.56THH219 pKa = 6.5FGGDD223 pKa = 3.79PVGVDD228 pKa = 3.35YY229 pKa = 11.03TSFSDD234 pKa = 3.45VNLRR238 pKa = 11.84EE239 pKa = 4.04EE240 pKa = 5.07FEE242 pKa = 4.17IEE244 pKa = 3.88QNIPRR249 pKa = 11.84SSTPANKK256 pKa = 10.23NIATKK261 pKa = 10.71AKK263 pKa = 10.37DD264 pKa = 3.14LYY266 pKa = 11.17QRR268 pKa = 11.84FTLQVPTQSLGAVARR283 pKa = 11.84PSRR286 pKa = 11.84QVTFQFEE293 pKa = 4.11NPAFSEE299 pKa = 4.33DD300 pKa = 2.94VTMAFNQDD308 pKa = 3.37VEE310 pKa = 4.71EE311 pKa = 4.36VTAAPNEE318 pKa = 4.06QFRR321 pKa = 11.84DD322 pKa = 3.4IQTLSRR328 pKa = 11.84PRR330 pKa = 11.84FSEE333 pKa = 4.17TEE335 pKa = 3.95SGHH338 pKa = 5.73VRR340 pKa = 11.84LSRR343 pKa = 11.84IGQKK347 pKa = 10.98ASMKK351 pKa = 9.17TRR353 pKa = 11.84SGLIIGEE360 pKa = 4.2RR361 pKa = 11.84VHH363 pKa = 6.15YY364 pKa = 9.69FYY366 pKa = 10.68DD367 pKa = 3.46ISDD370 pKa = 3.43ITVGEE375 pKa = 4.26NIEE378 pKa = 4.57LPTISSSVQTSGEE391 pKa = 4.28SIIQDD396 pKa = 3.57SQIDD400 pKa = 4.62GIVLDD405 pKa = 5.01EE406 pKa = 4.52NLLDD410 pKa = 5.54DD411 pKa = 4.48FAEE414 pKa = 4.66DD415 pKa = 4.05FSNAHH420 pKa = 7.11LIFTTTDD427 pKa = 3.1EE428 pKa = 4.55FNEE431 pKa = 4.25SVDD434 pKa = 3.69IIGIPPEE441 pKa = 4.33FGLKK445 pKa = 9.25TFIPQYY451 pKa = 9.89TNNSLDD457 pKa = 3.36SSQISMPVLPLSIPSAPLYY476 pKa = 8.74PAAVIIFGEE485 pKa = 4.55DD486 pKa = 3.69YY487 pKa = 10.8IIDD490 pKa = 4.12PYY492 pKa = 9.98YY493 pKa = 10.69LRR495 pKa = 11.84RR496 pKa = 11.84KK497 pKa = 9.16RR498 pKa = 11.84KK499 pKa = 9.65RR500 pKa = 11.84VDD502 pKa = 4.5FYY504 pKa = 11.93
MM1 pKa = 7.24SRR3 pKa = 11.84ARR5 pKa = 11.84RR6 pKa = 11.84TKK8 pKa = 9.73RR9 pKa = 11.84ASVQDD14 pKa = 4.7LYY16 pKa = 10.98TSCKK20 pKa = 10.4LGGDD24 pKa = 3.74CPEE27 pKa = 4.09DD28 pKa = 3.27VKK30 pKa = 11.65NKK32 pKa = 10.07VEE34 pKa = 4.07NTTLADD40 pKa = 3.79RR41 pKa = 11.84LLQILGSVVYY51 pKa = 9.79FGQLGIGSGKK61 pKa = 8.28GTGGITGYY69 pKa = 10.59RR70 pKa = 11.84PLQNTPRR77 pKa = 11.84VGTNRR82 pKa = 11.84PIEE85 pKa = 4.43SIRR88 pKa = 11.84PNIPIDD94 pKa = 3.64PLGGSDD100 pKa = 4.51IIPPTVIDD108 pKa = 3.41ATAPSVVPLTEE119 pKa = 3.96TGTPEE124 pKa = 3.68VNIISSGTGTNIEE137 pKa = 4.59LGEE140 pKa = 4.23LDD142 pKa = 4.39IITNVDD148 pKa = 3.44PTSTAVGTAEE158 pKa = 4.25HH159 pKa = 6.95PAVIDD164 pKa = 3.6VTDD167 pKa = 4.05EE168 pKa = 4.07GTSVIDD174 pKa = 3.94VQSGPPPPKK183 pKa = 9.87RR184 pKa = 11.84LQIDD188 pKa = 3.75SSLKK192 pKa = 8.95PYY194 pKa = 10.22EE195 pKa = 4.68HH196 pKa = 7.43IEE198 pKa = 4.03LTVYY202 pKa = 10.36PPYY205 pKa = 10.47NQSDD209 pKa = 3.86PNVNVFVDD217 pKa = 3.56THH219 pKa = 6.5FGGDD223 pKa = 3.79PVGVDD228 pKa = 3.35YY229 pKa = 11.03TSFSDD234 pKa = 3.45VNLRR238 pKa = 11.84EE239 pKa = 4.04EE240 pKa = 5.07FEE242 pKa = 4.17IEE244 pKa = 3.88QNIPRR249 pKa = 11.84SSTPANKK256 pKa = 10.23NIATKK261 pKa = 10.71AKK263 pKa = 10.37DD264 pKa = 3.14LYY266 pKa = 11.17QRR268 pKa = 11.84FTLQVPTQSLGAVARR283 pKa = 11.84PSRR286 pKa = 11.84QVTFQFEE293 pKa = 4.11NPAFSEE299 pKa = 4.33DD300 pKa = 2.94VTMAFNQDD308 pKa = 3.37VEE310 pKa = 4.71EE311 pKa = 4.36VTAAPNEE318 pKa = 4.06QFRR321 pKa = 11.84DD322 pKa = 3.4IQTLSRR328 pKa = 11.84PRR330 pKa = 11.84FSEE333 pKa = 4.17TEE335 pKa = 3.95SGHH338 pKa = 5.73VRR340 pKa = 11.84LSRR343 pKa = 11.84IGQKK347 pKa = 10.98ASMKK351 pKa = 9.17TRR353 pKa = 11.84SGLIIGEE360 pKa = 4.2RR361 pKa = 11.84VHH363 pKa = 6.15YY364 pKa = 9.69FYY366 pKa = 10.68DD367 pKa = 3.46ISDD370 pKa = 3.43ITVGEE375 pKa = 4.26NIEE378 pKa = 4.57LPTISSSVQTSGEE391 pKa = 4.28SIIQDD396 pKa = 3.57SQIDD400 pKa = 4.62GIVLDD405 pKa = 5.01EE406 pKa = 4.52NLLDD410 pKa = 5.54DD411 pKa = 4.48FAEE414 pKa = 4.66DD415 pKa = 4.05FSNAHH420 pKa = 7.11LIFTTTDD427 pKa = 3.1EE428 pKa = 4.55FNEE431 pKa = 4.25SVDD434 pKa = 3.69IIGIPPEE441 pKa = 4.33FGLKK445 pKa = 9.25TFIPQYY451 pKa = 9.89TNNSLDD457 pKa = 3.36SSQISMPVLPLSIPSAPLYY476 pKa = 8.74PAAVIIFGEE485 pKa = 4.55DD486 pKa = 3.69YY487 pKa = 10.8IIDD490 pKa = 4.12PYY492 pKa = 9.98YY493 pKa = 10.69LRR495 pKa = 11.84RR496 pKa = 11.84KK497 pKa = 9.16RR498 pKa = 11.84KK499 pKa = 9.65RR500 pKa = 11.84VDD502 pKa = 4.5FYY504 pKa = 11.93
Molecular weight: 55.28 kDa
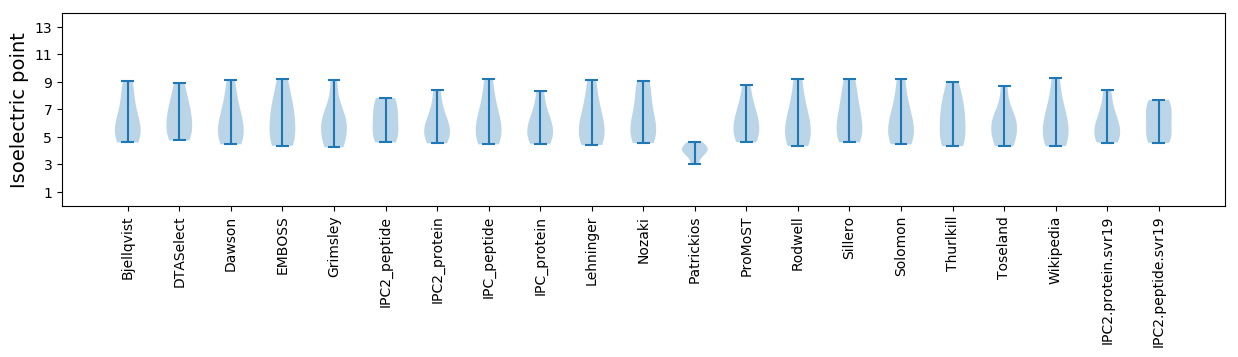
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2D2ALR4|A0A2D2ALR4_9PAPI Minor capsid protein L2 OS=Gammapapillomavirus 13 OX=1513258 GN=L2 PE=3 SV=1
MM1 pKa = 7.01NQADD5 pKa = 3.84LTRR8 pKa = 11.84RR9 pKa = 11.84FDD11 pKa = 3.67ALQDD15 pKa = 3.6EE16 pKa = 5.22LLTLIEE22 pKa = 4.94SGSDD26 pKa = 3.39DD27 pKa = 4.34LSTQIRR33 pKa = 11.84HH34 pKa = 5.47WEE36 pKa = 4.01LNRR39 pKa = 11.84KK40 pKa = 9.2INVTMYY46 pKa = 9.29YY47 pKa = 10.24GRR49 pKa = 11.84KK50 pKa = 9.09EE51 pKa = 4.02GYY53 pKa = 10.24KK54 pKa = 10.59NFGLQQLPAPQVSEE68 pKa = 4.4YY69 pKa = 10.03NAKK72 pKa = 10.25LAITMQILLKK82 pKa = 10.81SLATSQYY89 pKa = 10.71KK90 pKa = 9.9SEE92 pKa = 4.11PWSLTDD98 pKa = 3.11TSTEE102 pKa = 4.02VLLTPPKK109 pKa = 10.82NCFKK113 pKa = 11.11KK114 pKa = 10.45LGYY117 pKa = 7.91QVKK120 pKa = 7.97VTFDD124 pKa = 4.52DD125 pKa = 4.8DD126 pKa = 3.94PEE128 pKa = 5.89NYY130 pKa = 9.33MLYY133 pKa = 9.87TNWDD137 pKa = 3.62AIYY140 pKa = 10.54YY141 pKa = 10.01QDD143 pKa = 5.68LQDD146 pKa = 3.39IWHH149 pKa = 6.65KK150 pKa = 10.9VPGGVDD156 pKa = 3.51HH157 pKa = 7.22NGLYY161 pKa = 10.48YY162 pKa = 10.78DD163 pKa = 4.66DD164 pKa = 3.89VTGEE168 pKa = 3.96RR169 pKa = 11.84NYY171 pKa = 10.85FLLFQPEE178 pKa = 4.0AEE180 pKa = 4.45RR181 pKa = 11.84YY182 pKa = 7.7GKK184 pKa = 10.41SGIWTVQYY192 pKa = 11.37KK193 pKa = 8.4NTTLSSIVTSSSKK206 pKa = 10.67YY207 pKa = 9.77SSAGYY212 pKa = 8.75SAEE215 pKa = 3.97YY216 pKa = 8.98TADD219 pKa = 3.3SGGYY223 pKa = 9.26SSPEE227 pKa = 3.72EE228 pKa = 4.09GTSRR232 pKa = 11.84RR233 pKa = 11.84SQEE236 pKa = 3.53ASSRR240 pKa = 11.84SSGQPTSTSEE250 pKa = 3.47VGRR253 pKa = 11.84RR254 pKa = 11.84RR255 pKa = 11.84RR256 pKa = 11.84KK257 pKa = 9.09QGEE260 pKa = 4.25SGTEE264 pKa = 3.57RR265 pKa = 11.84STGGVSRR272 pKa = 11.84KK273 pKa = 7.85RR274 pKa = 11.84QRR276 pKa = 11.84PTSAPSPEE284 pKa = 4.31EE285 pKa = 3.41VGRR288 pKa = 11.84RR289 pKa = 11.84HH290 pKa = 5.81QSLPRR295 pKa = 11.84TGLSRR300 pKa = 11.84LRR302 pKa = 11.84RR303 pKa = 11.84LEE305 pKa = 3.89EE306 pKa = 3.83EE307 pKa = 4.38ARR309 pKa = 11.84DD310 pKa = 3.89PPLLLIKK317 pKa = 10.54GPPNTLKK324 pKa = 10.54CWRR327 pKa = 11.84FRR329 pKa = 11.84CNNRR333 pKa = 11.84CGHH336 pKa = 7.24LFTSISTVFKK346 pKa = 8.8WTSDD350 pKa = 3.6DD351 pKa = 3.67YY352 pKa = 11.96CNNPLQANGRR362 pKa = 11.84MLISFKK368 pKa = 10.79DD369 pKa = 3.35LQQRR373 pKa = 11.84QKK375 pKa = 11.02FITHH379 pKa = 5.21VNIPRR384 pKa = 11.84HH385 pKa = 4.54TSYY388 pKa = 11.85ALGSVDD394 pKa = 4.47SLL396 pKa = 3.95
MM1 pKa = 7.01NQADD5 pKa = 3.84LTRR8 pKa = 11.84RR9 pKa = 11.84FDD11 pKa = 3.67ALQDD15 pKa = 3.6EE16 pKa = 5.22LLTLIEE22 pKa = 4.94SGSDD26 pKa = 3.39DD27 pKa = 4.34LSTQIRR33 pKa = 11.84HH34 pKa = 5.47WEE36 pKa = 4.01LNRR39 pKa = 11.84KK40 pKa = 9.2INVTMYY46 pKa = 9.29YY47 pKa = 10.24GRR49 pKa = 11.84KK50 pKa = 9.09EE51 pKa = 4.02GYY53 pKa = 10.24KK54 pKa = 10.59NFGLQQLPAPQVSEE68 pKa = 4.4YY69 pKa = 10.03NAKK72 pKa = 10.25LAITMQILLKK82 pKa = 10.81SLATSQYY89 pKa = 10.71KK90 pKa = 9.9SEE92 pKa = 4.11PWSLTDD98 pKa = 3.11TSTEE102 pKa = 4.02VLLTPPKK109 pKa = 10.82NCFKK113 pKa = 11.11KK114 pKa = 10.45LGYY117 pKa = 7.91QVKK120 pKa = 7.97VTFDD124 pKa = 4.52DD125 pKa = 4.8DD126 pKa = 3.94PEE128 pKa = 5.89NYY130 pKa = 9.33MLYY133 pKa = 9.87TNWDD137 pKa = 3.62AIYY140 pKa = 10.54YY141 pKa = 10.01QDD143 pKa = 5.68LQDD146 pKa = 3.39IWHH149 pKa = 6.65KK150 pKa = 10.9VPGGVDD156 pKa = 3.51HH157 pKa = 7.22NGLYY161 pKa = 10.48YY162 pKa = 10.78DD163 pKa = 4.66DD164 pKa = 3.89VTGEE168 pKa = 3.96RR169 pKa = 11.84NYY171 pKa = 10.85FLLFQPEE178 pKa = 4.0AEE180 pKa = 4.45RR181 pKa = 11.84YY182 pKa = 7.7GKK184 pKa = 10.41SGIWTVQYY192 pKa = 11.37KK193 pKa = 8.4NTTLSSIVTSSSKK206 pKa = 10.67YY207 pKa = 9.77SSAGYY212 pKa = 8.75SAEE215 pKa = 3.97YY216 pKa = 8.98TADD219 pKa = 3.3SGGYY223 pKa = 9.26SSPEE227 pKa = 3.72EE228 pKa = 4.09GTSRR232 pKa = 11.84RR233 pKa = 11.84SQEE236 pKa = 3.53ASSRR240 pKa = 11.84SSGQPTSTSEE250 pKa = 3.47VGRR253 pKa = 11.84RR254 pKa = 11.84RR255 pKa = 11.84RR256 pKa = 11.84KK257 pKa = 9.09QGEE260 pKa = 4.25SGTEE264 pKa = 3.57RR265 pKa = 11.84STGGVSRR272 pKa = 11.84KK273 pKa = 7.85RR274 pKa = 11.84QRR276 pKa = 11.84PTSAPSPEE284 pKa = 4.31EE285 pKa = 3.41VGRR288 pKa = 11.84RR289 pKa = 11.84HH290 pKa = 5.81QSLPRR295 pKa = 11.84TGLSRR300 pKa = 11.84LRR302 pKa = 11.84RR303 pKa = 11.84LEE305 pKa = 3.89EE306 pKa = 3.83EE307 pKa = 4.38ARR309 pKa = 11.84DD310 pKa = 3.89PPLLLIKK317 pKa = 10.54GPPNTLKK324 pKa = 10.54CWRR327 pKa = 11.84FRR329 pKa = 11.84CNNRR333 pKa = 11.84CGHH336 pKa = 7.24LFTSISTVFKK346 pKa = 8.8WTSDD350 pKa = 3.6DD351 pKa = 3.67YY352 pKa = 11.96CNNPLQANGRR362 pKa = 11.84MLISFKK368 pKa = 10.79DD369 pKa = 3.35LQQRR373 pKa = 11.84QKK375 pKa = 11.02FITHH379 pKa = 5.21VNIPRR384 pKa = 11.84HH385 pKa = 4.54TSYY388 pKa = 11.85ALGSVDD394 pKa = 4.47SLL396 pKa = 3.95
Molecular weight: 45.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2383 |
96 |
603 |
340.4 |
38.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.952 ± 0.52 | 2.14 ± 0.7 |
7.092 ± 0.439 | 5.791 ± 0.339 |
4.784 ± 0.597 | 5.455 ± 0.573 |
1.804 ± 0.271 | 6.253 ± 0.871 |
5.287 ± 0.783 | 8.687 ± 0.56 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.385 ± 0.274 | 5.371 ± 0.511 |
6.085 ± 0.841 | 4.658 ± 0.251 |
5.749 ± 0.611 | 7.637 ± 1.123 |
6.42 ± 0.627 | 5.791 ± 0.605 |
1.175 ± 0.324 | 3.483 ± 0.451 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
