
Porphyridium purpureum (Red alga) (Porphyridium cruentum)
Taxonomy: cellular organisms; Eukaryota; Rhodophyta; Bangiophyceae; Porphyridiales; Porphyridiaceae; Porphyridium
Average proteome isoelectric point is 6.82
Get precalculated fractions of proteins
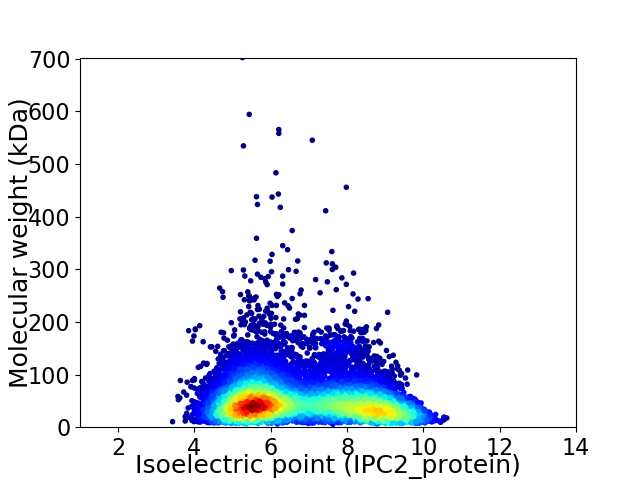
Virtual 2D-PAGE plot for 9673 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5J4YMY5|A0A5J4YMY5_PORPP Uncharacterized protein OS=Porphyridium purpureum OX=35688 GN=FVE85_8505 PE=4 SV=1
MM1 pKa = 8.07LMFLGCVAAVPWVNQLPLDD20 pKa = 4.04EE21 pKa = 5.29TSTACCVSSFTVDD34 pKa = 3.1AEE36 pKa = 4.41GFGNVEE42 pKa = 4.22LVASVGMTDD51 pKa = 3.52GTAGVPEE58 pKa = 4.23ATNQGGIDD66 pKa = 3.33MVIRR70 pKa = 11.84VMGAPFRR77 pKa = 11.84GDD79 pKa = 3.0GDD81 pKa = 3.51VGGQQTSLGSILIGTDD97 pKa = 3.33GEE99 pKa = 5.03DD100 pKa = 2.8IATNVAFDD108 pKa = 3.6AQANQLVVIGTTTGNLAGGDD128 pKa = 4.06GFPQSQRR135 pKa = 11.84DD136 pKa = 3.56LFMAFIDD143 pKa = 3.83VTVEE147 pKa = 3.79NPNQVTLAVNTVVQCGPGEE166 pKa = 4.26GTNTPFALLRR176 pKa = 11.84LPGSVRR182 pKa = 11.84WLGLANSAATEE193 pKa = 4.01NGPFPTGLGFIRR205 pKa = 11.84SDD207 pKa = 4.06DD208 pKa = 3.71VCNGNNFQQKK218 pKa = 9.88NGNTDD223 pKa = 3.51GYY225 pKa = 10.66FSGYY229 pKa = 9.6AAAVKK234 pKa = 9.71TGEE237 pKa = 4.25DD238 pKa = 3.25VFAYY242 pKa = 10.11VYY244 pKa = 10.39EE245 pKa = 4.46VATNPSGGQVYY256 pKa = 9.63RR257 pKa = 11.84AEE259 pKa = 4.08QYY261 pKa = 10.86VLQDD265 pKa = 2.84GDD267 pKa = 3.84FNEE270 pKa = 4.36EE271 pKa = 3.55FRR273 pKa = 11.84IDD275 pKa = 4.85LEE277 pKa = 4.3FDD279 pKa = 3.51PLGQGGLAFEE289 pKa = 4.96SSSITYY295 pKa = 8.1DD296 pKa = 3.12TDD298 pKa = 3.2NSLFALAGVRR308 pKa = 11.84PGYY311 pKa = 11.01LNTEE315 pKa = 3.68HH316 pKa = 7.56DD317 pKa = 3.73AAEE320 pKa = 4.48TEE322 pKa = 4.31VFTYY326 pKa = 10.4YY327 pKa = 11.18AATFTDD333 pKa = 4.82DD334 pKa = 3.13SDD336 pKa = 5.5LNVEE340 pKa = 4.32VAGDD344 pKa = 3.84STDD347 pKa = 3.17GGTVIPVQPVVISAGGSAYY366 pKa = 10.19AAYY369 pKa = 8.6TKK371 pKa = 10.29SRR373 pKa = 11.84RR374 pKa = 11.84DD375 pKa = 3.41RR376 pKa = 11.84VPWAGRR382 pKa = 11.84IFDD385 pKa = 4.56LPSLSFNEE393 pKa = 4.1GDD395 pKa = 4.15DD396 pKa = 3.83FLSVGNLTDD405 pKa = 5.81DD406 pKa = 3.43IVNQIVGFIAPRR418 pKa = 11.84TSQTSFSGFYY428 pKa = 10.38YY429 pKa = 10.9GMAFTPEE436 pKa = 4.28EE437 pKa = 4.15NPFNCVGGPDD447 pKa = 3.28RR448 pKa = 11.84KK449 pKa = 10.41DD450 pKa = 2.99FAMAGLGFSGDD461 pKa = 3.62NVSEE465 pKa = 4.09SWIFTVPSQCSEE477 pKa = 4.25VAVDD481 pKa = 4.8AVLQASVSSDD491 pKa = 2.86RR492 pKa = 11.84PIFVTGTTTGSIAGTTQPTNGGATTTQWTMRR523 pKa = 11.84IEE525 pKa = 4.65PGTGQLPPLNPLCDD539 pKa = 4.62LPCQSIPPPTPSPTPSPTPSPTLSPSPTPDD569 pKa = 3.27ANQTASPSPSPSPSPTPTPTIGSLPVSDD597 pKa = 4.61SVDD600 pKa = 3.17GGVIAGAVVGAIAGSALIVGIGAWIYY626 pKa = 10.71KK627 pKa = 9.51YY628 pKa = 10.75KK629 pKa = 10.45SDD631 pKa = 3.48QAAEE635 pKa = 4.38GGFGEE640 pKa = 4.61TFEE643 pKa = 4.34
MM1 pKa = 8.07LMFLGCVAAVPWVNQLPLDD20 pKa = 4.04EE21 pKa = 5.29TSTACCVSSFTVDD34 pKa = 3.1AEE36 pKa = 4.41GFGNVEE42 pKa = 4.22LVASVGMTDD51 pKa = 3.52GTAGVPEE58 pKa = 4.23ATNQGGIDD66 pKa = 3.33MVIRR70 pKa = 11.84VMGAPFRR77 pKa = 11.84GDD79 pKa = 3.0GDD81 pKa = 3.51VGGQQTSLGSILIGTDD97 pKa = 3.33GEE99 pKa = 5.03DD100 pKa = 2.8IATNVAFDD108 pKa = 3.6AQANQLVVIGTTTGNLAGGDD128 pKa = 4.06GFPQSQRR135 pKa = 11.84DD136 pKa = 3.56LFMAFIDD143 pKa = 3.83VTVEE147 pKa = 3.79NPNQVTLAVNTVVQCGPGEE166 pKa = 4.26GTNTPFALLRR176 pKa = 11.84LPGSVRR182 pKa = 11.84WLGLANSAATEE193 pKa = 4.01NGPFPTGLGFIRR205 pKa = 11.84SDD207 pKa = 4.06DD208 pKa = 3.71VCNGNNFQQKK218 pKa = 9.88NGNTDD223 pKa = 3.51GYY225 pKa = 10.66FSGYY229 pKa = 9.6AAAVKK234 pKa = 9.71TGEE237 pKa = 4.25DD238 pKa = 3.25VFAYY242 pKa = 10.11VYY244 pKa = 10.39EE245 pKa = 4.46VATNPSGGQVYY256 pKa = 9.63RR257 pKa = 11.84AEE259 pKa = 4.08QYY261 pKa = 10.86VLQDD265 pKa = 2.84GDD267 pKa = 3.84FNEE270 pKa = 4.36EE271 pKa = 3.55FRR273 pKa = 11.84IDD275 pKa = 4.85LEE277 pKa = 4.3FDD279 pKa = 3.51PLGQGGLAFEE289 pKa = 4.96SSSITYY295 pKa = 8.1DD296 pKa = 3.12TDD298 pKa = 3.2NSLFALAGVRR308 pKa = 11.84PGYY311 pKa = 11.01LNTEE315 pKa = 3.68HH316 pKa = 7.56DD317 pKa = 3.73AAEE320 pKa = 4.48TEE322 pKa = 4.31VFTYY326 pKa = 10.4YY327 pKa = 11.18AATFTDD333 pKa = 4.82DD334 pKa = 3.13SDD336 pKa = 5.5LNVEE340 pKa = 4.32VAGDD344 pKa = 3.84STDD347 pKa = 3.17GGTVIPVQPVVISAGGSAYY366 pKa = 10.19AAYY369 pKa = 8.6TKK371 pKa = 10.29SRR373 pKa = 11.84RR374 pKa = 11.84DD375 pKa = 3.41RR376 pKa = 11.84VPWAGRR382 pKa = 11.84IFDD385 pKa = 4.56LPSLSFNEE393 pKa = 4.1GDD395 pKa = 4.15DD396 pKa = 3.83FLSVGNLTDD405 pKa = 5.81DD406 pKa = 3.43IVNQIVGFIAPRR418 pKa = 11.84TSQTSFSGFYY428 pKa = 10.38YY429 pKa = 10.9GMAFTPEE436 pKa = 4.28EE437 pKa = 4.15NPFNCVGGPDD447 pKa = 3.28RR448 pKa = 11.84KK449 pKa = 10.41DD450 pKa = 2.99FAMAGLGFSGDD461 pKa = 3.62NVSEE465 pKa = 4.09SWIFTVPSQCSEE477 pKa = 4.25VAVDD481 pKa = 4.8AVLQASVSSDD491 pKa = 2.86RR492 pKa = 11.84PIFVTGTTTGSIAGTTQPTNGGATTTQWTMRR523 pKa = 11.84IEE525 pKa = 4.65PGTGQLPPLNPLCDD539 pKa = 4.62LPCQSIPPPTPSPTPSPTPSPTLSPSPTPDD569 pKa = 3.27ANQTASPSPSPSPSPTPTPTIGSLPVSDD597 pKa = 4.61SVDD600 pKa = 3.17GGVIAGAVVGAIAGSALIVGIGAWIYY626 pKa = 10.71KK627 pKa = 9.51YY628 pKa = 10.75KK629 pKa = 10.45SDD631 pKa = 3.48QAAEE635 pKa = 4.38GGFGEE640 pKa = 4.61TFEE643 pKa = 4.34
Molecular weight: 66.79 kDa
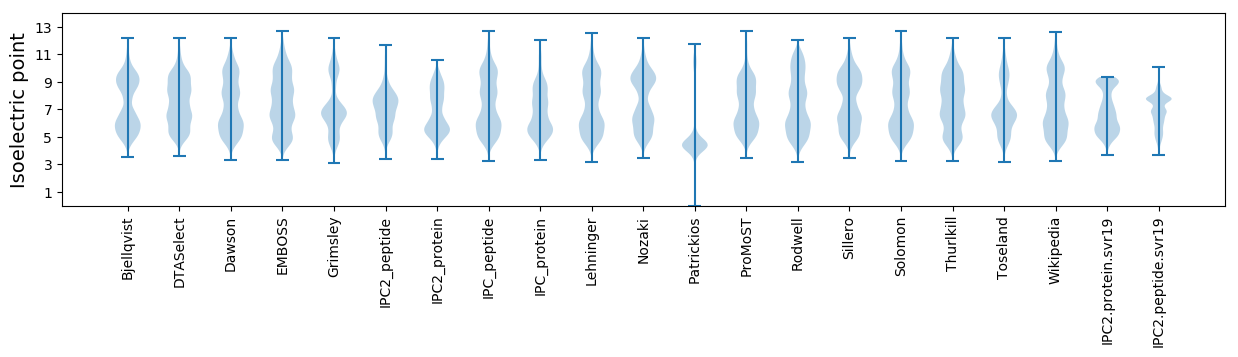
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5J4Z810|A0A5J4Z810_PORPP ATP-dependent RNA helicase eIF4A OS=Porphyridium purpureum OX=35688 GN=FVE85_6825 PE=3 SV=1
MM1 pKa = 7.26FGWVHH6 pKa = 5.93SVPPRR11 pKa = 11.84LQPHH15 pKa = 4.68QTPSFGNVCAGRR27 pKa = 11.84FARR30 pKa = 11.84FDD32 pKa = 3.56VPNGQTRR39 pKa = 11.84ARR41 pKa = 11.84ARR43 pKa = 11.84ARR45 pKa = 11.84TAPEE49 pKa = 3.99LPPRR53 pKa = 11.84WTRR56 pKa = 11.84TTVRR60 pKa = 11.84RR61 pKa = 11.84KK62 pKa = 9.77DD63 pKa = 3.24AAVPRR68 pKa = 11.84TEE70 pKa = 4.11QFPFPVMNEE79 pKa = 3.63ALKK82 pKa = 10.35QVPTAKK88 pKa = 10.59AMAVLLCIDD97 pKa = 4.48QIFKK101 pKa = 10.89NAFARR106 pKa = 11.84YY107 pKa = 7.9NWCFPSALAAMCALLGALLMLDD129 pKa = 3.32RR130 pKa = 11.84VSPDD134 pKa = 2.78SAEE137 pKa = 3.96NVVGFFQPVCAFFSRR152 pKa = 11.84WLAVFFVPVLIALPVSARR170 pKa = 11.84PWPAADD176 pKa = 3.83EE177 pKa = 4.15LAKK180 pKa = 10.44IAVVLVGGLAISLGFTAKK198 pKa = 10.39LCALLRR204 pKa = 11.84AASAGAEE211 pKa = 4.07KK212 pKa = 10.54DD213 pKa = 3.48GQEE216 pKa = 4.13QVSLAGAASVPLSTGAPSASATSFPCANLTRR247 pKa = 11.84FWGGVALLSFCSGVMFPAVQPLRR270 pKa = 11.84LVFTFATTLFGFCMGQRR287 pKa = 11.84AAPSVKK293 pKa = 10.02RR294 pKa = 11.84LVHH297 pKa = 5.92PLVATVLTSFLAFSLFGRR315 pKa = 11.84LSGIGFFNVVGEE327 pKa = 4.19YY328 pKa = 10.12FSKK331 pKa = 10.75GKK333 pKa = 10.55CGACLGGGDD342 pKa = 5.04LLMMILGPAIVSFAFHH358 pKa = 6.53IFNRR362 pKa = 11.84RR363 pKa = 11.84ALIKK367 pKa = 9.47RR368 pKa = 11.84HH369 pKa = 5.31APEE372 pKa = 4.23VVGTTALSSFVSLFATAYY390 pKa = 9.79VGRR393 pKa = 11.84LLNLPANIRR402 pKa = 11.84LSLIPRR408 pKa = 11.84MVTAPLAICIGDD420 pKa = 4.63LLHH423 pKa = 7.49ADD425 pKa = 3.76PALAGALAATTGLLGANFYY444 pKa = 11.85AMVLDD449 pKa = 4.7GLQITDD455 pKa = 4.81PIYY458 pKa = 10.62RR459 pKa = 11.84GISTGSASHH468 pKa = 7.01GIGSAAILKK477 pKa = 9.73EE478 pKa = 3.95GEE480 pKa = 4.19PFAFAAIAMVLCGVFSTVFISIKK503 pKa = 8.98PLRR506 pKa = 11.84AALIARR512 pKa = 11.84RR513 pKa = 11.84MTGHH517 pKa = 7.63PLNSIPRR524 pKa = 11.84LLPRR528 pKa = 11.84SLPGTPASTRR538 pKa = 11.84KK539 pKa = 9.57QSLAIAPSPDD549 pKa = 3.0VRR551 pKa = 11.84CRR553 pKa = 11.84TNNRR557 pKa = 11.84GSGRR561 pKa = 11.84VNEE564 pKa = 4.47VGGAMRR570 pKa = 11.84FRR572 pKa = 11.84CRR574 pKa = 11.84SLVVSPRR581 pKa = 11.84CAYY584 pKa = 9.73IPSNSNFSSS593 pKa = 3.32
MM1 pKa = 7.26FGWVHH6 pKa = 5.93SVPPRR11 pKa = 11.84LQPHH15 pKa = 4.68QTPSFGNVCAGRR27 pKa = 11.84FARR30 pKa = 11.84FDD32 pKa = 3.56VPNGQTRR39 pKa = 11.84ARR41 pKa = 11.84ARR43 pKa = 11.84ARR45 pKa = 11.84TAPEE49 pKa = 3.99LPPRR53 pKa = 11.84WTRR56 pKa = 11.84TTVRR60 pKa = 11.84RR61 pKa = 11.84KK62 pKa = 9.77DD63 pKa = 3.24AAVPRR68 pKa = 11.84TEE70 pKa = 4.11QFPFPVMNEE79 pKa = 3.63ALKK82 pKa = 10.35QVPTAKK88 pKa = 10.59AMAVLLCIDD97 pKa = 4.48QIFKK101 pKa = 10.89NAFARR106 pKa = 11.84YY107 pKa = 7.9NWCFPSALAAMCALLGALLMLDD129 pKa = 3.32RR130 pKa = 11.84VSPDD134 pKa = 2.78SAEE137 pKa = 3.96NVVGFFQPVCAFFSRR152 pKa = 11.84WLAVFFVPVLIALPVSARR170 pKa = 11.84PWPAADD176 pKa = 3.83EE177 pKa = 4.15LAKK180 pKa = 10.44IAVVLVGGLAISLGFTAKK198 pKa = 10.39LCALLRR204 pKa = 11.84AASAGAEE211 pKa = 4.07KK212 pKa = 10.54DD213 pKa = 3.48GQEE216 pKa = 4.13QVSLAGAASVPLSTGAPSASATSFPCANLTRR247 pKa = 11.84FWGGVALLSFCSGVMFPAVQPLRR270 pKa = 11.84LVFTFATTLFGFCMGQRR287 pKa = 11.84AAPSVKK293 pKa = 10.02RR294 pKa = 11.84LVHH297 pKa = 5.92PLVATVLTSFLAFSLFGRR315 pKa = 11.84LSGIGFFNVVGEE327 pKa = 4.19YY328 pKa = 10.12FSKK331 pKa = 10.75GKK333 pKa = 10.55CGACLGGGDD342 pKa = 5.04LLMMILGPAIVSFAFHH358 pKa = 6.53IFNRR362 pKa = 11.84RR363 pKa = 11.84ALIKK367 pKa = 9.47RR368 pKa = 11.84HH369 pKa = 5.31APEE372 pKa = 4.23VVGTTALSSFVSLFATAYY390 pKa = 9.79VGRR393 pKa = 11.84LLNLPANIRR402 pKa = 11.84LSLIPRR408 pKa = 11.84MVTAPLAICIGDD420 pKa = 4.63LLHH423 pKa = 7.49ADD425 pKa = 3.76PALAGALAATTGLLGANFYY444 pKa = 11.85AMVLDD449 pKa = 4.7GLQITDD455 pKa = 4.81PIYY458 pKa = 10.62RR459 pKa = 11.84GISTGSASHH468 pKa = 7.01GIGSAAILKK477 pKa = 9.73EE478 pKa = 3.95GEE480 pKa = 4.19PFAFAAIAMVLCGVFSTVFISIKK503 pKa = 8.98PLRR506 pKa = 11.84AALIARR512 pKa = 11.84RR513 pKa = 11.84MTGHH517 pKa = 7.63PLNSIPRR524 pKa = 11.84LLPRR528 pKa = 11.84SLPGTPASTRR538 pKa = 11.84KK539 pKa = 9.57QSLAIAPSPDD549 pKa = 3.0VRR551 pKa = 11.84CRR553 pKa = 11.84TNNRR557 pKa = 11.84GSGRR561 pKa = 11.84VNEE564 pKa = 4.47VGGAMRR570 pKa = 11.84FRR572 pKa = 11.84CRR574 pKa = 11.84SLVVSPRR581 pKa = 11.84CAYY584 pKa = 9.73IPSNSNFSSS593 pKa = 3.32
Molecular weight: 63.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4998014 |
50 |
6419 |
516.7 |
56.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.5 ± 0.025 | 1.693 ± 0.009 |
5.538 ± 0.015 | 6.402 ± 0.019 |
3.605 ± 0.015 | 6.964 ± 0.021 |
2.537 ± 0.012 | 3.645 ± 0.013 |
4.234 ± 0.022 | 9.368 ± 0.024 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.54 ± 0.009 | 2.927 ± 0.011 |
4.803 ± 0.016 | 4.033 ± 0.016 |
7.393 ± 0.022 | 8.124 ± 0.025 |
4.849 ± 0.012 | 7.356 ± 0.017 |
1.273 ± 0.008 | 2.215 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
