
Wenzhou sobemo-like virus 4
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.73
Get precalculated fractions of proteins
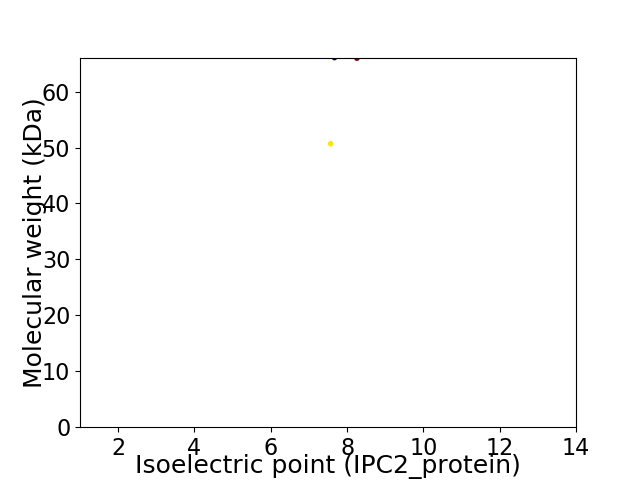
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEG7|A0A1L3KEG7_9VIRU RNA-directed RNA polymerase OS=Wenzhou sobemo-like virus 4 OX=1923660 PE=4 SV=1
MM1 pKa = 7.63PANGLHH7 pKa = 5.95VPDD10 pKa = 4.42TFGEE14 pKa = 4.22LPYY17 pKa = 10.71GIPQVDD23 pKa = 3.19ACSAKK28 pKa = 9.87RR29 pKa = 11.84SFRR32 pKa = 11.84AHH34 pKa = 5.89CQKK37 pKa = 10.52QKK39 pKa = 9.2QTDD42 pKa = 3.69APPTAEE48 pKa = 3.7EE49 pKa = 3.98RR50 pKa = 11.84TRR52 pKa = 11.84AVQTALRR59 pKa = 11.84SYY61 pKa = 9.84SAWRR65 pKa = 11.84FDD67 pKa = 3.73LTSAADD73 pKa = 3.56PNAWFDD79 pKa = 4.27SEE81 pKa = 4.42FNKK84 pKa = 10.48ALKK87 pKa = 10.01EE88 pKa = 3.51IDD90 pKa = 3.6MNSTPGYY97 pKa = 11.06CMLTNLGSNNAQIFGWDD114 pKa = 3.71GLTFDD119 pKa = 5.12PDD121 pKa = 3.13RR122 pKa = 11.84VSLVRR127 pKa = 11.84QHH129 pKa = 5.49VKK131 pKa = 10.53LRR133 pKa = 11.84FDD135 pKa = 3.34EE136 pKa = 4.63LLYY139 pKa = 11.4GNAVCDD145 pKa = 4.57DD146 pKa = 3.2IKK148 pKa = 11.45VFVKK152 pKa = 10.13QEE154 pKa = 3.55PHH156 pKa = 6.28KK157 pKa = 10.69LKK159 pKa = 10.53KK160 pKa = 9.98IQDD163 pKa = 3.36GTYY166 pKa = 9.79RR167 pKa = 11.84LISAVSLIDD176 pKa = 3.52TLIDD180 pKa = 4.32RR181 pKa = 11.84ILFAWIARR189 pKa = 11.84AQLSTVGEE197 pKa = 4.63TPCLIGWSPVRR208 pKa = 11.84GGWRR212 pKa = 11.84LITNKK217 pKa = 9.65FANEE221 pKa = 3.78PVNCLDD227 pKa = 3.68RR228 pKa = 11.84SAWDD232 pKa = 3.36WTVQGYY238 pKa = 8.9LVDD241 pKa = 3.93LWILFLEE248 pKa = 4.37NLPVNPPEE256 pKa = 3.71WWLKK260 pKa = 8.27MMKK263 pKa = 10.07LRR265 pKa = 11.84FKK267 pKa = 11.1LLFEE271 pKa = 4.65DD272 pKa = 3.89AVFRR276 pKa = 11.84FEE278 pKa = 5.04DD279 pKa = 3.83GTRR282 pKa = 11.84VRR284 pKa = 11.84QGTKK288 pKa = 9.93GVMKK292 pKa = 10.06SGCYY296 pKa = 8.47LTILLNSLSQSLLHH310 pKa = 6.08YY311 pKa = 9.88IANKK315 pKa = 9.43RR316 pKa = 11.84CGRR319 pKa = 11.84PMALKK324 pKa = 10.34QPFSIGDD331 pKa = 3.59DD332 pKa = 3.71TVQEE336 pKa = 4.1AMSWLPEE343 pKa = 3.85YY344 pKa = 10.66VRR346 pKa = 11.84QLEE349 pKa = 4.15ILGITVKK356 pKa = 10.34GAKK359 pKa = 7.74VQHH362 pKa = 5.26WVEE365 pKa = 3.88FAGFCFDD372 pKa = 6.05GKK374 pKa = 8.79TCYY377 pKa = 8.86PAYY380 pKa = 8.17WQKK383 pKa = 11.1HH384 pKa = 4.58LFNLKK389 pKa = 8.15HH390 pKa = 4.81TQRR393 pKa = 11.84LGEE396 pKa = 4.09TLMSYY401 pKa = 10.41QYY403 pKa = 11.1LYY405 pKa = 11.1VNEE408 pKa = 3.83PVMYY412 pKa = 10.47EE413 pKa = 3.89FVCRR417 pKa = 11.84VARR420 pKa = 11.84EE421 pKa = 3.85LGPRR425 pKa = 11.84NVLPRR430 pKa = 11.84IEE432 pKa = 5.34ALDD435 pKa = 3.55IMNQPRR441 pKa = 3.26
MM1 pKa = 7.63PANGLHH7 pKa = 5.95VPDD10 pKa = 4.42TFGEE14 pKa = 4.22LPYY17 pKa = 10.71GIPQVDD23 pKa = 3.19ACSAKK28 pKa = 9.87RR29 pKa = 11.84SFRR32 pKa = 11.84AHH34 pKa = 5.89CQKK37 pKa = 10.52QKK39 pKa = 9.2QTDD42 pKa = 3.69APPTAEE48 pKa = 3.7EE49 pKa = 3.98RR50 pKa = 11.84TRR52 pKa = 11.84AVQTALRR59 pKa = 11.84SYY61 pKa = 9.84SAWRR65 pKa = 11.84FDD67 pKa = 3.73LTSAADD73 pKa = 3.56PNAWFDD79 pKa = 4.27SEE81 pKa = 4.42FNKK84 pKa = 10.48ALKK87 pKa = 10.01EE88 pKa = 3.51IDD90 pKa = 3.6MNSTPGYY97 pKa = 11.06CMLTNLGSNNAQIFGWDD114 pKa = 3.71GLTFDD119 pKa = 5.12PDD121 pKa = 3.13RR122 pKa = 11.84VSLVRR127 pKa = 11.84QHH129 pKa = 5.49VKK131 pKa = 10.53LRR133 pKa = 11.84FDD135 pKa = 3.34EE136 pKa = 4.63LLYY139 pKa = 11.4GNAVCDD145 pKa = 4.57DD146 pKa = 3.2IKK148 pKa = 11.45VFVKK152 pKa = 10.13QEE154 pKa = 3.55PHH156 pKa = 6.28KK157 pKa = 10.69LKK159 pKa = 10.53KK160 pKa = 9.98IQDD163 pKa = 3.36GTYY166 pKa = 9.79RR167 pKa = 11.84LISAVSLIDD176 pKa = 3.52TLIDD180 pKa = 4.32RR181 pKa = 11.84ILFAWIARR189 pKa = 11.84AQLSTVGEE197 pKa = 4.63TPCLIGWSPVRR208 pKa = 11.84GGWRR212 pKa = 11.84LITNKK217 pKa = 9.65FANEE221 pKa = 3.78PVNCLDD227 pKa = 3.68RR228 pKa = 11.84SAWDD232 pKa = 3.36WTVQGYY238 pKa = 8.9LVDD241 pKa = 3.93LWILFLEE248 pKa = 4.37NLPVNPPEE256 pKa = 3.71WWLKK260 pKa = 8.27MMKK263 pKa = 10.07LRR265 pKa = 11.84FKK267 pKa = 11.1LLFEE271 pKa = 4.65DD272 pKa = 3.89AVFRR276 pKa = 11.84FEE278 pKa = 5.04DD279 pKa = 3.83GTRR282 pKa = 11.84VRR284 pKa = 11.84QGTKK288 pKa = 9.93GVMKK292 pKa = 10.06SGCYY296 pKa = 8.47LTILLNSLSQSLLHH310 pKa = 6.08YY311 pKa = 9.88IANKK315 pKa = 9.43RR316 pKa = 11.84CGRR319 pKa = 11.84PMALKK324 pKa = 10.34QPFSIGDD331 pKa = 3.59DD332 pKa = 3.71TVQEE336 pKa = 4.1AMSWLPEE343 pKa = 3.85YY344 pKa = 10.66VRR346 pKa = 11.84QLEE349 pKa = 4.15ILGITVKK356 pKa = 10.34GAKK359 pKa = 7.74VQHH362 pKa = 5.26WVEE365 pKa = 3.88FAGFCFDD372 pKa = 6.05GKK374 pKa = 8.79TCYY377 pKa = 8.86PAYY380 pKa = 8.17WQKK383 pKa = 11.1HH384 pKa = 4.58LFNLKK389 pKa = 8.15HH390 pKa = 4.81TQRR393 pKa = 11.84LGEE396 pKa = 4.09TLMSYY401 pKa = 10.41QYY403 pKa = 11.1LYY405 pKa = 11.1VNEE408 pKa = 3.83PVMYY412 pKa = 10.47EE413 pKa = 3.89FVCRR417 pKa = 11.84VARR420 pKa = 11.84EE421 pKa = 3.85LGPRR425 pKa = 11.84NVLPRR430 pKa = 11.84IEE432 pKa = 5.34ALDD435 pKa = 3.55IMNQPRR441 pKa = 3.26
Molecular weight: 50.68 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEG7|A0A1L3KEG7_9VIRU RNA-directed RNA polymerase OS=Wenzhou sobemo-like virus 4 OX=1923660 PE=4 SV=1
MM1 pKa = 7.53TIYY4 pKa = 10.93ANVMEE9 pKa = 4.72ILYY12 pKa = 10.74SSVIYY17 pKa = 9.35VQIVAMVILAITALNVVNPLNKK39 pKa = 9.92TMILVSFGTIVAVLLRR55 pKa = 11.84SILYY59 pKa = 9.15WIEE62 pKa = 3.65TMVIMTYY69 pKa = 8.93GTAKK73 pKa = 10.28KK74 pKa = 9.76IWVEE78 pKa = 4.18LIFQRR83 pKa = 11.84FVNNLGTDD91 pKa = 3.54PNKK94 pKa = 10.11EE95 pKa = 3.99LKK97 pKa = 10.71LLIVMALIIIFMYY110 pKa = 9.18IWKK113 pKa = 9.09VCKK116 pKa = 10.04YY117 pKa = 8.93LQSTKK122 pKa = 9.98VVYY125 pKa = 9.53VVPSKK130 pKa = 10.87VKK132 pKa = 10.62KK133 pKa = 10.61GDD135 pKa = 3.1ACYY138 pKa = 10.46VEE140 pKa = 4.96EE141 pKa = 4.05RR142 pKa = 11.84AMPNSIYY149 pKa = 10.65EE150 pKa = 4.38HH151 pKa = 6.55IKK153 pKa = 10.39VLPPFQAMVMASLDD167 pKa = 3.63GYY169 pKa = 9.56KK170 pKa = 10.09YY171 pKa = 10.61HH172 pKa = 8.33VMGQCFWVDD181 pKa = 3.29EE182 pKa = 4.46GLVTAAHH189 pKa = 5.9VIEE192 pKa = 5.14GFEE195 pKa = 4.13HH196 pKa = 6.27LCIYY200 pKa = 10.5RR201 pKa = 11.84DD202 pKa = 3.8LEE204 pKa = 3.8HH205 pKa = 7.18SINIKK210 pKa = 9.18ATEE213 pKa = 4.25FEE215 pKa = 4.9CGNGDD220 pKa = 3.56YY221 pKa = 10.67AVCRR225 pKa = 11.84EE226 pKa = 4.22PTKK229 pKa = 9.83ITQKK233 pKa = 10.69VGLSKK238 pKa = 11.14AKK240 pKa = 10.02LSPVAVQKK248 pKa = 11.21NGGVTVNVSAMDD260 pKa = 3.31KK261 pKa = 9.59RR262 pKa = 11.84TAGFLDD268 pKa = 3.73EE269 pKa = 4.27HH270 pKa = 5.68TQFGFVTYY278 pKa = 9.83TGSTTKK284 pKa = 10.42GFSGAPYY291 pKa = 9.59CFGRR295 pKa = 11.84TVYY298 pKa = 10.77GMHH301 pKa = 7.16LGSDD305 pKa = 4.44SKK307 pKa = 12.08NMGYY311 pKa = 10.69DD312 pKa = 3.41GAFLKK317 pKa = 11.14AEE319 pKa = 4.72LKK321 pKa = 9.94PSRR324 pKa = 11.84LIKK327 pKa = 10.31QKK329 pKa = 11.0LGIKK333 pKa = 9.41TEE335 pKa = 5.2DD336 pKa = 3.34SAEE339 pKa = 3.98WLIGQVDD346 pKa = 4.48KK347 pKa = 11.38YY348 pKa = 11.69AEE350 pKa = 4.25LDD352 pKa = 3.78YY353 pKa = 11.29VRR355 pKa = 11.84SPGNPDD361 pKa = 2.59EE362 pKa = 4.0WRR364 pKa = 11.84VRR366 pKa = 11.84VGGRR370 pKa = 11.84YY371 pKa = 9.23HH372 pKa = 7.13LVDD375 pKa = 4.62DD376 pKa = 4.61EE377 pKa = 5.23VMATLLQARR386 pKa = 11.84KK387 pKa = 8.16GTEE390 pKa = 3.6VRR392 pKa = 11.84IGKK395 pKa = 9.75LKK397 pKa = 10.66DD398 pKa = 3.54FQRR401 pKa = 11.84EE402 pKa = 3.97ALFSRR407 pKa = 11.84GEE409 pKa = 3.85ISKK412 pKa = 10.64KK413 pKa = 10.47SSNHH417 pKa = 5.35PKK419 pKa = 9.9LAKK422 pKa = 10.63EE423 pKa = 4.0EE424 pKa = 4.26DD425 pKa = 3.69PEE427 pKa = 4.1KK428 pKa = 11.47AMVKK432 pKa = 10.42GLLAEE437 pKa = 5.05FGRR440 pKa = 11.84TSIVTEE446 pKa = 3.86EE447 pKa = 4.0PMVRR451 pKa = 11.84EE452 pKa = 4.31SGPSVDD458 pKa = 4.89GLPLAPRR465 pKa = 11.84DD466 pKa = 3.49AMSFNDD472 pKa = 3.53SGNLLRR478 pKa = 11.84APTVDD483 pKa = 3.29VGALGVEE490 pKa = 4.1NVRR493 pKa = 11.84RR494 pKa = 11.84EE495 pKa = 4.12FVRR498 pKa = 11.84NPSLQTCQQTVCTCQTPSVSYY519 pKa = 10.72HH520 pKa = 4.6MEE522 pKa = 3.86SRR524 pKa = 11.84KK525 pKa = 10.5SMPAQQSEE533 pKa = 3.91ASARR537 pKa = 11.84TVKK540 pKa = 10.66NKK542 pKa = 8.62NRR544 pKa = 11.84RR545 pKa = 11.84MLRR548 pKa = 11.84QQQKK552 pKa = 10.54KK553 pKa = 8.68EE554 pKa = 3.91LEE556 pKa = 4.32LYY558 pKa = 10.07KK559 pKa = 10.2QLCGPIQHH567 pKa = 6.79GASTLHH573 pKa = 5.78PQQTQMHH580 pKa = 7.16GSTQSLTKK588 pKa = 10.35HH589 pKa = 5.71
MM1 pKa = 7.53TIYY4 pKa = 10.93ANVMEE9 pKa = 4.72ILYY12 pKa = 10.74SSVIYY17 pKa = 9.35VQIVAMVILAITALNVVNPLNKK39 pKa = 9.92TMILVSFGTIVAVLLRR55 pKa = 11.84SILYY59 pKa = 9.15WIEE62 pKa = 3.65TMVIMTYY69 pKa = 8.93GTAKK73 pKa = 10.28KK74 pKa = 9.76IWVEE78 pKa = 4.18LIFQRR83 pKa = 11.84FVNNLGTDD91 pKa = 3.54PNKK94 pKa = 10.11EE95 pKa = 3.99LKK97 pKa = 10.71LLIVMALIIIFMYY110 pKa = 9.18IWKK113 pKa = 9.09VCKK116 pKa = 10.04YY117 pKa = 8.93LQSTKK122 pKa = 9.98VVYY125 pKa = 9.53VVPSKK130 pKa = 10.87VKK132 pKa = 10.62KK133 pKa = 10.61GDD135 pKa = 3.1ACYY138 pKa = 10.46VEE140 pKa = 4.96EE141 pKa = 4.05RR142 pKa = 11.84AMPNSIYY149 pKa = 10.65EE150 pKa = 4.38HH151 pKa = 6.55IKK153 pKa = 10.39VLPPFQAMVMASLDD167 pKa = 3.63GYY169 pKa = 9.56KK170 pKa = 10.09YY171 pKa = 10.61HH172 pKa = 8.33VMGQCFWVDD181 pKa = 3.29EE182 pKa = 4.46GLVTAAHH189 pKa = 5.9VIEE192 pKa = 5.14GFEE195 pKa = 4.13HH196 pKa = 6.27LCIYY200 pKa = 10.5RR201 pKa = 11.84DD202 pKa = 3.8LEE204 pKa = 3.8HH205 pKa = 7.18SINIKK210 pKa = 9.18ATEE213 pKa = 4.25FEE215 pKa = 4.9CGNGDD220 pKa = 3.56YY221 pKa = 10.67AVCRR225 pKa = 11.84EE226 pKa = 4.22PTKK229 pKa = 9.83ITQKK233 pKa = 10.69VGLSKK238 pKa = 11.14AKK240 pKa = 10.02LSPVAVQKK248 pKa = 11.21NGGVTVNVSAMDD260 pKa = 3.31KK261 pKa = 9.59RR262 pKa = 11.84TAGFLDD268 pKa = 3.73EE269 pKa = 4.27HH270 pKa = 5.68TQFGFVTYY278 pKa = 9.83TGSTTKK284 pKa = 10.42GFSGAPYY291 pKa = 9.59CFGRR295 pKa = 11.84TVYY298 pKa = 10.77GMHH301 pKa = 7.16LGSDD305 pKa = 4.44SKK307 pKa = 12.08NMGYY311 pKa = 10.69DD312 pKa = 3.41GAFLKK317 pKa = 11.14AEE319 pKa = 4.72LKK321 pKa = 9.94PSRR324 pKa = 11.84LIKK327 pKa = 10.31QKK329 pKa = 11.0LGIKK333 pKa = 9.41TEE335 pKa = 5.2DD336 pKa = 3.34SAEE339 pKa = 3.98WLIGQVDD346 pKa = 4.48KK347 pKa = 11.38YY348 pKa = 11.69AEE350 pKa = 4.25LDD352 pKa = 3.78YY353 pKa = 11.29VRR355 pKa = 11.84SPGNPDD361 pKa = 2.59EE362 pKa = 4.0WRR364 pKa = 11.84VRR366 pKa = 11.84VGGRR370 pKa = 11.84YY371 pKa = 9.23HH372 pKa = 7.13LVDD375 pKa = 4.62DD376 pKa = 4.61EE377 pKa = 5.23VMATLLQARR386 pKa = 11.84KK387 pKa = 8.16GTEE390 pKa = 3.6VRR392 pKa = 11.84IGKK395 pKa = 9.75LKK397 pKa = 10.66DD398 pKa = 3.54FQRR401 pKa = 11.84EE402 pKa = 3.97ALFSRR407 pKa = 11.84GEE409 pKa = 3.85ISKK412 pKa = 10.64KK413 pKa = 10.47SSNHH417 pKa = 5.35PKK419 pKa = 9.9LAKK422 pKa = 10.63EE423 pKa = 4.0EE424 pKa = 4.26DD425 pKa = 3.69PEE427 pKa = 4.1KK428 pKa = 11.47AMVKK432 pKa = 10.42GLLAEE437 pKa = 5.05FGRR440 pKa = 11.84TSIVTEE446 pKa = 3.86EE447 pKa = 4.0PMVRR451 pKa = 11.84EE452 pKa = 4.31SGPSVDD458 pKa = 4.89GLPLAPRR465 pKa = 11.84DD466 pKa = 3.49AMSFNDD472 pKa = 3.53SGNLLRR478 pKa = 11.84APTVDD483 pKa = 3.29VGALGVEE490 pKa = 4.1NVRR493 pKa = 11.84RR494 pKa = 11.84EE495 pKa = 4.12FVRR498 pKa = 11.84NPSLQTCQQTVCTCQTPSVSYY519 pKa = 10.72HH520 pKa = 4.6MEE522 pKa = 3.86SRR524 pKa = 11.84KK525 pKa = 10.5SMPAQQSEE533 pKa = 3.91ASARR537 pKa = 11.84TVKK540 pKa = 10.66NKK542 pKa = 8.62NRR544 pKa = 11.84RR545 pKa = 11.84MLRR548 pKa = 11.84QQQKK552 pKa = 10.54KK553 pKa = 8.68EE554 pKa = 3.91LEE556 pKa = 4.32LYY558 pKa = 10.07KK559 pKa = 10.2QLCGPIQHH567 pKa = 6.79GASTLHH573 pKa = 5.78PQQTQMHH580 pKa = 7.16GSTQSLTKK588 pKa = 10.35HH589 pKa = 5.71
Molecular weight: 65.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1030 |
441 |
589 |
515.0 |
58.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.602 ± 0.122 | 2.136 ± 0.217 |
4.563 ± 0.67 | 5.728 ± 0.448 |
3.981 ± 0.611 | 6.505 ± 0.369 |
2.136 ± 0.195 | 5.049 ± 0.311 |
6.602 ± 0.703 | 9.515 ± 0.83 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.301 ± 0.489 | 3.883 ± 0.257 |
4.66 ± 0.474 | 4.66 ± 0.062 |
5.437 ± 0.553 | 5.728 ± 0.585 |
5.728 ± 0.311 | 8.155 ± 0.957 |
1.942 ± 0.747 | 3.689 ± 0.174 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
