
Sclerotinia sclerotiorum hypovirulence associated DNA virus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus sclero1; Sclerotinia gemycircularvirus 1
Average proteome isoelectric point is 7.47
Get precalculated fractions of proteins
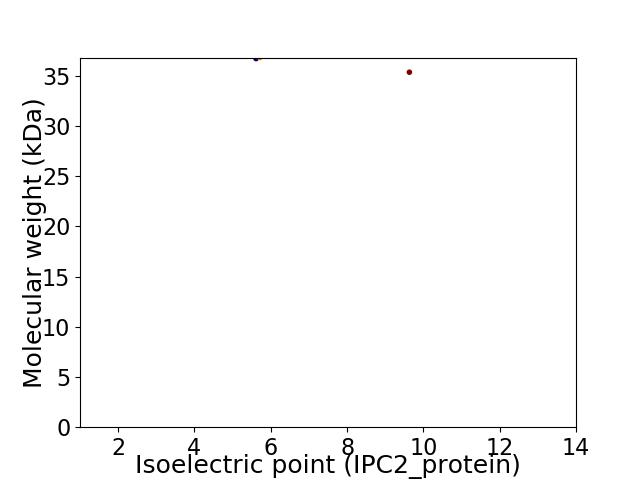
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C7FFR1|C7FFR1_9VIRU Replication-associated protein OS=Sclerotinia sclerotiorum hypovirulence associated DNA virus 1 OX=664785 GN=AC1 PE=3 SV=1
MM1 pKa = 7.42TFDD4 pKa = 3.38FHH6 pKa = 8.1AKK8 pKa = 9.84YY9 pKa = 10.92VLLTYY14 pKa = 9.04AQCGEE19 pKa = 3.98LDD21 pKa = 3.5AFRR24 pKa = 11.84VMDD27 pKa = 4.21KK28 pKa = 11.15LSLLGAEE35 pKa = 4.59CIIGRR40 pKa = 11.84EE41 pKa = 4.01HH42 pKa = 7.75HH43 pKa = 6.83EE44 pKa = 4.72DD45 pKa = 3.5GGTHH49 pKa = 5.81LHH51 pKa = 6.57CFAEE55 pKa = 5.16FGRR58 pKa = 11.84KK59 pKa = 8.16FRR61 pKa = 11.84SRR63 pKa = 11.84KK64 pKa = 9.78ADD66 pKa = 3.3VFDD69 pKa = 4.55VDD71 pKa = 3.74GHH73 pKa = 6.53HH74 pKa = 7.33PNITKK79 pKa = 10.39SRR81 pKa = 11.84GTPEE85 pKa = 3.56KK86 pKa = 10.7GYY88 pKa = 10.89DD89 pKa = 3.47YY90 pKa = 10.69AIKK93 pKa = 10.71DD94 pKa = 3.36GDD96 pKa = 4.38VICGGLGRR104 pKa = 11.84PSVGRR109 pKa = 11.84VGTRR113 pKa = 11.84PSDD116 pKa = 3.87SKK118 pKa = 10.61WAIITSASNRR128 pKa = 11.84DD129 pKa = 3.71EE130 pKa = 4.26FWEE133 pKa = 4.4LVHH136 pKa = 7.01EE137 pKa = 5.14LDD139 pKa = 4.19PKK141 pKa = 10.74AAATSFSQLQRR152 pKa = 11.84YY153 pKa = 8.74CDD155 pKa = 3.02WKK157 pKa = 9.32YY158 pKa = 10.58QYY160 pKa = 10.22HH161 pKa = 5.55APEE164 pKa = 4.41YY165 pKa = 8.76EE166 pKa = 4.57SPAGAHH172 pKa = 6.75FIGAEE177 pKa = 3.59LDD179 pKa = 3.47GRR181 pKa = 11.84NRR183 pKa = 11.84WLEE186 pKa = 3.56QSGIGSSEE194 pKa = 3.79EE195 pKa = 3.33RR196 pKa = 11.84RR197 pKa = 11.84GRR199 pKa = 11.84VKK201 pKa = 10.81SLVLYY206 pKa = 10.59GPSQTGKK213 pKa = 8.63TSWARR218 pKa = 11.84SLGKK222 pKa = 9.77HH223 pKa = 6.1IYY225 pKa = 10.15CVGLVSGTEE234 pKa = 4.02CLKK237 pKa = 11.26APDD240 pKa = 3.26VEE242 pKa = 4.5YY243 pKa = 10.97AVFDD247 pKa = 5.14DD248 pKa = 3.44IRR250 pKa = 11.84GGIKK254 pKa = 10.03FFHH257 pKa = 6.71SFKK260 pKa = 10.56EE261 pKa = 3.93WLGCQPHH268 pKa = 6.79LSVKK272 pKa = 9.79EE273 pKa = 3.98LYY275 pKa = 10.11RR276 pKa = 11.84EE277 pKa = 3.96PKK279 pKa = 9.33VIEE282 pKa = 3.96WGKK285 pKa = 9.65PSIWCSNADD294 pKa = 3.54PRR296 pKa = 11.84NEE298 pKa = 3.86LLQVDD303 pKa = 4.54IDD305 pKa = 3.82WMEE308 pKa = 4.05MNCTFIEE315 pKa = 3.87ITEE318 pKa = 4.18PVVEE322 pKa = 4.81FNN324 pKa = 4.55
MM1 pKa = 7.42TFDD4 pKa = 3.38FHH6 pKa = 8.1AKK8 pKa = 9.84YY9 pKa = 10.92VLLTYY14 pKa = 9.04AQCGEE19 pKa = 3.98LDD21 pKa = 3.5AFRR24 pKa = 11.84VMDD27 pKa = 4.21KK28 pKa = 11.15LSLLGAEE35 pKa = 4.59CIIGRR40 pKa = 11.84EE41 pKa = 4.01HH42 pKa = 7.75HH43 pKa = 6.83EE44 pKa = 4.72DD45 pKa = 3.5GGTHH49 pKa = 5.81LHH51 pKa = 6.57CFAEE55 pKa = 5.16FGRR58 pKa = 11.84KK59 pKa = 8.16FRR61 pKa = 11.84SRR63 pKa = 11.84KK64 pKa = 9.78ADD66 pKa = 3.3VFDD69 pKa = 4.55VDD71 pKa = 3.74GHH73 pKa = 6.53HH74 pKa = 7.33PNITKK79 pKa = 10.39SRR81 pKa = 11.84GTPEE85 pKa = 3.56KK86 pKa = 10.7GYY88 pKa = 10.89DD89 pKa = 3.47YY90 pKa = 10.69AIKK93 pKa = 10.71DD94 pKa = 3.36GDD96 pKa = 4.38VICGGLGRR104 pKa = 11.84PSVGRR109 pKa = 11.84VGTRR113 pKa = 11.84PSDD116 pKa = 3.87SKK118 pKa = 10.61WAIITSASNRR128 pKa = 11.84DD129 pKa = 3.71EE130 pKa = 4.26FWEE133 pKa = 4.4LVHH136 pKa = 7.01EE137 pKa = 5.14LDD139 pKa = 4.19PKK141 pKa = 10.74AAATSFSQLQRR152 pKa = 11.84YY153 pKa = 8.74CDD155 pKa = 3.02WKK157 pKa = 9.32YY158 pKa = 10.58QYY160 pKa = 10.22HH161 pKa = 5.55APEE164 pKa = 4.41YY165 pKa = 8.76EE166 pKa = 4.57SPAGAHH172 pKa = 6.75FIGAEE177 pKa = 3.59LDD179 pKa = 3.47GRR181 pKa = 11.84NRR183 pKa = 11.84WLEE186 pKa = 3.56QSGIGSSEE194 pKa = 3.79EE195 pKa = 3.33RR196 pKa = 11.84RR197 pKa = 11.84GRR199 pKa = 11.84VKK201 pKa = 10.81SLVLYY206 pKa = 10.59GPSQTGKK213 pKa = 8.63TSWARR218 pKa = 11.84SLGKK222 pKa = 9.77HH223 pKa = 6.1IYY225 pKa = 10.15CVGLVSGTEE234 pKa = 4.02CLKK237 pKa = 11.26APDD240 pKa = 3.26VEE242 pKa = 4.5YY243 pKa = 10.97AVFDD247 pKa = 5.14DD248 pKa = 3.44IRR250 pKa = 11.84GGIKK254 pKa = 10.03FFHH257 pKa = 6.71SFKK260 pKa = 10.56EE261 pKa = 3.93WLGCQPHH268 pKa = 6.79LSVKK272 pKa = 9.79EE273 pKa = 3.98LYY275 pKa = 10.11RR276 pKa = 11.84EE277 pKa = 3.96PKK279 pKa = 9.33VIEE282 pKa = 3.96WGKK285 pKa = 9.65PSIWCSNADD294 pKa = 3.54PRR296 pKa = 11.84NEE298 pKa = 3.86LLQVDD303 pKa = 4.54IDD305 pKa = 3.82WMEE308 pKa = 4.05MNCTFIEE315 pKa = 3.87ITEE318 pKa = 4.18PVVEE322 pKa = 4.81FNN324 pKa = 4.55
Molecular weight: 36.72 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C7FFR1|C7FFR1_9VIRU Replication-associated protein OS=Sclerotinia sclerotiorum hypovirulence associated DNA virus 1 OX=664785 GN=AC1 PE=3 SV=1
MM1 pKa = 7.83AYY3 pKa = 10.26ARR5 pKa = 11.84YY6 pKa = 9.19RR7 pKa = 11.84SSRR10 pKa = 11.84FRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84PIKK18 pKa = 7.98TSRR21 pKa = 11.84RR22 pKa = 11.84GGVSKK27 pKa = 10.3PRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84SYY33 pKa = 10.12SAKK36 pKa = 9.66KK37 pKa = 8.24RR38 pKa = 11.84TYY40 pKa = 9.61RR41 pKa = 11.84KK42 pKa = 8.9KK43 pKa = 10.36ASMSKK48 pKa = 10.34RR49 pKa = 11.84SILNTTSRR57 pKa = 11.84KK58 pKa = 9.22KK59 pKa = 10.51RR60 pKa = 11.84NGMLTFSNTTNSGASTAPAAAPVYY84 pKa = 10.39VNAVLGGTFIWCPTAMNLVAQSLISNMSSHH114 pKa = 6.65TATTCYY120 pKa = 9.61MRR122 pKa = 11.84GLSEE126 pKa = 5.06HH127 pKa = 6.59IRR129 pKa = 11.84IQTSSGIPWFHH140 pKa = 6.81RR141 pKa = 11.84RR142 pKa = 11.84VCFTWKK148 pKa = 10.48GLGPFNVMATEE159 pKa = 4.68SPTSDD164 pKa = 4.33FPWLPYY170 pKa = 9.71QDD172 pKa = 3.82SSNGVQRR179 pKa = 11.84LFFNEE184 pKa = 3.94NVNNMPQTINRR195 pKa = 11.84QRR197 pKa = 11.84NILFKK202 pKa = 10.77GAEE205 pKa = 4.22NVDD208 pKa = 3.07WTDD211 pKa = 4.24PIIAPLDD218 pKa = 3.41TGRR221 pKa = 11.84ITVKK225 pKa = 10.33FDD227 pKa = 3.36KK228 pKa = 10.0TWTMQSGNTNGIVRR242 pKa = 11.84EE243 pKa = 4.01RR244 pKa = 11.84KK245 pKa = 8.63LWHH248 pKa = 6.56PMNHH252 pKa = 5.87NLVYY256 pKa = 10.89DD257 pKa = 4.1DD258 pKa = 5.94DD259 pKa = 4.85EE260 pKa = 6.18YY261 pKa = 11.99GDD263 pKa = 4.17IEE265 pKa = 4.49SGSWFSTDD273 pKa = 2.8SKK275 pKa = 11.63AGMGDD280 pKa = 4.16YY281 pKa = 11.03YY282 pKa = 11.64VMDD285 pKa = 5.83IIQGGQGGTASDD297 pKa = 4.69LLRR300 pKa = 11.84MTANSTMYY308 pKa = 9.24WHH310 pKa = 7.07EE311 pKa = 4.05KK312 pKa = 9.5
MM1 pKa = 7.83AYY3 pKa = 10.26ARR5 pKa = 11.84YY6 pKa = 9.19RR7 pKa = 11.84SSRR10 pKa = 11.84FRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84PIKK18 pKa = 7.98TSRR21 pKa = 11.84RR22 pKa = 11.84GGVSKK27 pKa = 10.3PRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84SYY33 pKa = 10.12SAKK36 pKa = 9.66KK37 pKa = 8.24RR38 pKa = 11.84TYY40 pKa = 9.61RR41 pKa = 11.84KK42 pKa = 8.9KK43 pKa = 10.36ASMSKK48 pKa = 10.34RR49 pKa = 11.84SILNTTSRR57 pKa = 11.84KK58 pKa = 9.22KK59 pKa = 10.51RR60 pKa = 11.84NGMLTFSNTTNSGASTAPAAAPVYY84 pKa = 10.39VNAVLGGTFIWCPTAMNLVAQSLISNMSSHH114 pKa = 6.65TATTCYY120 pKa = 9.61MRR122 pKa = 11.84GLSEE126 pKa = 5.06HH127 pKa = 6.59IRR129 pKa = 11.84IQTSSGIPWFHH140 pKa = 6.81RR141 pKa = 11.84RR142 pKa = 11.84VCFTWKK148 pKa = 10.48GLGPFNVMATEE159 pKa = 4.68SPTSDD164 pKa = 4.33FPWLPYY170 pKa = 9.71QDD172 pKa = 3.82SSNGVQRR179 pKa = 11.84LFFNEE184 pKa = 3.94NVNNMPQTINRR195 pKa = 11.84QRR197 pKa = 11.84NILFKK202 pKa = 10.77GAEE205 pKa = 4.22NVDD208 pKa = 3.07WTDD211 pKa = 4.24PIIAPLDD218 pKa = 3.41TGRR221 pKa = 11.84ITVKK225 pKa = 10.33FDD227 pKa = 3.36KK228 pKa = 10.0TWTMQSGNTNGIVRR242 pKa = 11.84EE243 pKa = 4.01RR244 pKa = 11.84KK245 pKa = 8.63LWHH248 pKa = 6.56PMNHH252 pKa = 5.87NLVYY256 pKa = 10.89DD257 pKa = 4.1DD258 pKa = 5.94DD259 pKa = 4.85EE260 pKa = 6.18YY261 pKa = 11.99GDD263 pKa = 4.17IEE265 pKa = 4.49SGSWFSTDD273 pKa = 2.8SKK275 pKa = 11.63AGMGDD280 pKa = 4.16YY281 pKa = 11.03YY282 pKa = 11.64VMDD285 pKa = 5.83IIQGGQGGTASDD297 pKa = 4.69LLRR300 pKa = 11.84MTANSTMYY308 pKa = 9.24WHH310 pKa = 7.07EE311 pKa = 4.05KK312 pKa = 9.5
Molecular weight: 35.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
636 |
312 |
324 |
318.0 |
36.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.289 ± 0.084 | 2.044 ± 0.75 |
5.503 ± 0.703 | 5.346 ± 1.926 |
4.403 ± 0.385 | 8.333 ± 0.666 |
2.987 ± 0.737 | 5.189 ± 0.042 |
5.503 ± 0.26 | 5.975 ± 0.808 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.83 ± 1.147 | 4.403 ± 1.612 |
4.403 ± 0.059 | 2.673 ± 0.147 |
7.39 ± 0.875 | 8.491 ± 1.001 |
6.447 ± 1.75 | 5.189 ± 0.486 |
2.83 ± 0.038 | 3.774 ± 0.05 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
