
Lake Sarah-associated circular virus-43
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 8.06
Get precalculated fractions of proteins
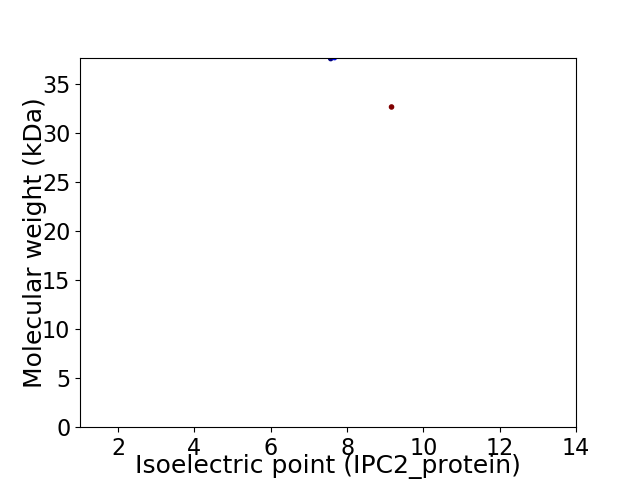
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
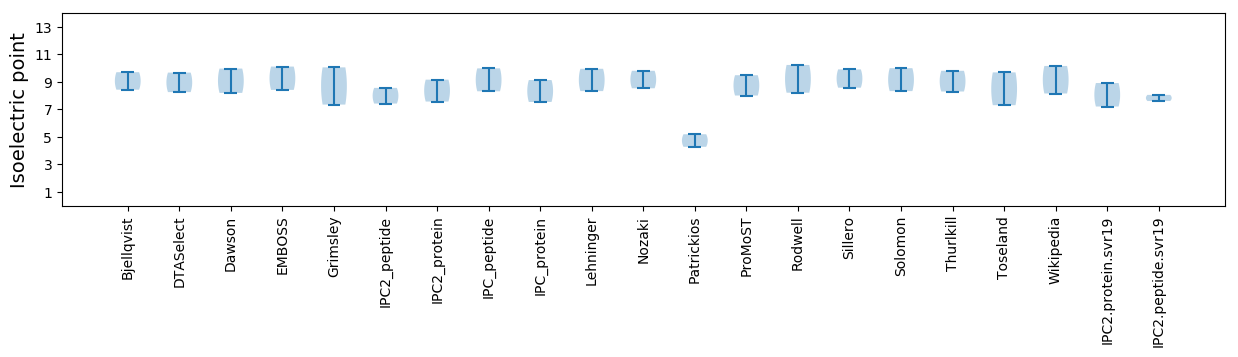
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A126GAE9|A0A126GAE9_9VIRU Putative capsid protein OS=Lake Sarah-associated circular virus-43 OX=1685772 PE=4 SV=1
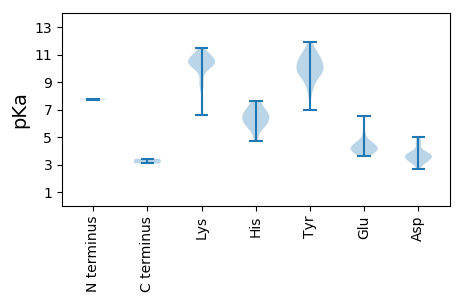
MM1 pKa = 7.73QSRR4 pKa = 11.84CWCFTVNFKK13 pKa = 10.78YY14 pKa = 11.0LNDD17 pKa = 3.72YY18 pKa = 9.95EE19 pKa = 6.56AIDD22 pKa = 3.68PAEE25 pKa = 3.96WPGCVVAVWQHH36 pKa = 4.74EE37 pKa = 4.38VGEE40 pKa = 4.77GGTEE44 pKa = 3.85HH45 pKa = 6.57LQGYY49 pKa = 8.92VNFAKK54 pKa = 10.18PKK56 pKa = 10.48RR57 pKa = 11.84MGALHH62 pKa = 6.82KK63 pKa = 10.54LSGMARR69 pKa = 11.84ASVRR73 pKa = 11.84PRR75 pKa = 11.84AKK77 pKa = 10.59LATKK81 pKa = 10.14KK82 pKa = 10.81DD83 pKa = 3.4NLVYY87 pKa = 9.42CTKK90 pKa = 10.89EE91 pKa = 3.77EE92 pKa = 4.1GRR94 pKa = 11.84LDD96 pKa = 3.6GPFYY100 pKa = 10.7FPSKK104 pKa = 10.23EE105 pKa = 4.02KK106 pKa = 11.12VEE108 pKa = 4.57AYY110 pKa = 10.31CKK112 pKa = 10.55VEE114 pKa = 3.62NGQRR118 pKa = 11.84TDD120 pKa = 3.32LALISDD126 pKa = 3.77QVARR130 pKa = 11.84GLTDD134 pKa = 3.74KK135 pKa = 10.71EE136 pKa = 3.79IAEE139 pKa = 4.37INPIAILKK147 pKa = 6.63YY148 pKa = 10.35QKK150 pKa = 10.88GIDD153 pKa = 3.71HH154 pKa = 6.8LRR156 pKa = 11.84IAITDD161 pKa = 3.67TSRR164 pKa = 11.84LGDD167 pKa = 3.78EE168 pKa = 4.26VDD170 pKa = 3.38SMVFVGPTGTGKK182 pKa = 10.25SYY184 pKa = 11.05RR185 pKa = 11.84LNRR188 pKa = 11.84DD189 pKa = 3.25YY190 pKa = 11.4PEE192 pKa = 4.56GPEE195 pKa = 4.14WFWVSPGKK203 pKa = 9.83WFDD206 pKa = 3.69GYY208 pKa = 10.77QGQPGLVFDD217 pKa = 4.5EE218 pKa = 6.03FRR220 pKa = 11.84DD221 pKa = 3.2NWYY224 pKa = 9.35PYY226 pKa = 10.7QFLLRR231 pKa = 11.84LLDD234 pKa = 3.67TKK236 pKa = 10.4PYY238 pKa = 9.97RR239 pKa = 11.84VEE241 pKa = 3.91KK242 pKa = 10.69KK243 pKa = 10.41GGHH246 pKa = 5.98LSMRR250 pKa = 11.84AHH252 pKa = 7.65RR253 pKa = 11.84FRR255 pKa = 11.84FSTNIHH261 pKa = 5.82PMHH264 pKa = 7.46WYY266 pKa = 9.43QGVVKK271 pKa = 8.86PAWEE275 pKa = 3.9EE276 pKa = 4.17SPLKK280 pKa = 10.45RR281 pKa = 11.84RR282 pKa = 11.84LPYY285 pKa = 9.82IEE287 pKa = 5.27LMEE290 pKa = 4.23VPYY293 pKa = 10.68DD294 pKa = 3.72RR295 pKa = 11.84PTVSFDD301 pKa = 3.88SAAAWALLQPEE312 pKa = 4.61AVAPAARR319 pKa = 11.84AVYY322 pKa = 8.37GQRR325 pKa = 11.84MDD327 pKa = 3.2GG328 pKa = 3.41
MM1 pKa = 7.73QSRR4 pKa = 11.84CWCFTVNFKK13 pKa = 10.78YY14 pKa = 11.0LNDD17 pKa = 3.72YY18 pKa = 9.95EE19 pKa = 6.56AIDD22 pKa = 3.68PAEE25 pKa = 3.96WPGCVVAVWQHH36 pKa = 4.74EE37 pKa = 4.38VGEE40 pKa = 4.77GGTEE44 pKa = 3.85HH45 pKa = 6.57LQGYY49 pKa = 8.92VNFAKK54 pKa = 10.18PKK56 pKa = 10.48RR57 pKa = 11.84MGALHH62 pKa = 6.82KK63 pKa = 10.54LSGMARR69 pKa = 11.84ASVRR73 pKa = 11.84PRR75 pKa = 11.84AKK77 pKa = 10.59LATKK81 pKa = 10.14KK82 pKa = 10.81DD83 pKa = 3.4NLVYY87 pKa = 9.42CTKK90 pKa = 10.89EE91 pKa = 3.77EE92 pKa = 4.1GRR94 pKa = 11.84LDD96 pKa = 3.6GPFYY100 pKa = 10.7FPSKK104 pKa = 10.23EE105 pKa = 4.02KK106 pKa = 11.12VEE108 pKa = 4.57AYY110 pKa = 10.31CKK112 pKa = 10.55VEE114 pKa = 3.62NGQRR118 pKa = 11.84TDD120 pKa = 3.32LALISDD126 pKa = 3.77QVARR130 pKa = 11.84GLTDD134 pKa = 3.74KK135 pKa = 10.71EE136 pKa = 3.79IAEE139 pKa = 4.37INPIAILKK147 pKa = 6.63YY148 pKa = 10.35QKK150 pKa = 10.88GIDD153 pKa = 3.71HH154 pKa = 6.8LRR156 pKa = 11.84IAITDD161 pKa = 3.67TSRR164 pKa = 11.84LGDD167 pKa = 3.78EE168 pKa = 4.26VDD170 pKa = 3.38SMVFVGPTGTGKK182 pKa = 10.25SYY184 pKa = 11.05RR185 pKa = 11.84LNRR188 pKa = 11.84DD189 pKa = 3.25YY190 pKa = 11.4PEE192 pKa = 4.56GPEE195 pKa = 4.14WFWVSPGKK203 pKa = 9.83WFDD206 pKa = 3.69GYY208 pKa = 10.77QGQPGLVFDD217 pKa = 4.5EE218 pKa = 6.03FRR220 pKa = 11.84DD221 pKa = 3.2NWYY224 pKa = 9.35PYY226 pKa = 10.7QFLLRR231 pKa = 11.84LLDD234 pKa = 3.67TKK236 pKa = 10.4PYY238 pKa = 9.97RR239 pKa = 11.84VEE241 pKa = 3.91KK242 pKa = 10.69KK243 pKa = 10.41GGHH246 pKa = 5.98LSMRR250 pKa = 11.84AHH252 pKa = 7.65RR253 pKa = 11.84FRR255 pKa = 11.84FSTNIHH261 pKa = 5.82PMHH264 pKa = 7.46WYY266 pKa = 9.43QGVVKK271 pKa = 8.86PAWEE275 pKa = 3.9EE276 pKa = 4.17SPLKK280 pKa = 10.45RR281 pKa = 11.84RR282 pKa = 11.84LPYY285 pKa = 9.82IEE287 pKa = 5.27LMEE290 pKa = 4.23VPYY293 pKa = 10.68DD294 pKa = 3.72RR295 pKa = 11.84PTVSFDD301 pKa = 3.88SAAAWALLQPEE312 pKa = 4.61AVAPAARR319 pKa = 11.84AVYY322 pKa = 8.37GQRR325 pKa = 11.84MDD327 pKa = 3.2GG328 pKa = 3.41
Molecular weight: 37.61 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A126GAE9|A0A126GAE9_9VIRU Putative capsid protein OS=Lake Sarah-associated circular virus-43 OX=1685772 PE=4 SV=1
MM1 pKa = 7.79PEE3 pKa = 4.12KK4 pKa = 10.39FLADD8 pKa = 4.74GISFHH13 pKa = 5.83HH14 pKa = 6.59HH15 pKa = 6.65KK16 pKa = 10.71KK17 pKa = 10.35LLCCAYY23 pKa = 9.68ILGINRR29 pKa = 11.84AKK31 pKa = 10.64GLRR34 pKa = 11.84IMSMYY39 pKa = 9.75YY40 pKa = 9.85RR41 pKa = 11.84KK42 pKa = 10.22KK43 pKa = 8.89KK44 pKa = 8.71TGGGGAARR52 pKa = 11.84GSVKK56 pKa = 10.42GYY58 pKa = 9.84AAAATRR64 pKa = 11.84VPGTAGKK71 pKa = 10.38AKK73 pKa = 10.26LAAARR78 pKa = 11.84NASKK82 pKa = 10.18SAQMARR88 pKa = 11.84YY89 pKa = 8.52VGRR92 pKa = 11.84RR93 pKa = 11.84SNVPGKK99 pKa = 10.08DD100 pKa = 2.7NHH102 pKa = 6.33TFDD105 pKa = 4.72PVVLNTNYY113 pKa = 10.07LQCDD117 pKa = 3.3NATAVAVSASGYY129 pKa = 6.97ITPGSSAIVLNQVSQGTSSTQRR151 pKa = 11.84IGRR154 pKa = 11.84KK155 pKa = 8.87ILMTAIHH162 pKa = 5.96IRR164 pKa = 11.84AYY166 pKa = 10.67LRR168 pKa = 11.84NTTASADD175 pKa = 3.61LNMVRR180 pKa = 11.84VALVYY185 pKa = 10.18IPEE188 pKa = 4.18MDD190 pKa = 3.25RR191 pKa = 11.84TTTTMPPQNAIWTSQDD207 pKa = 2.72ARR209 pKa = 11.84EE210 pKa = 4.5LRR212 pKa = 11.84VIDD215 pKa = 3.37NADD218 pKa = 3.49RR219 pKa = 11.84FKK221 pKa = 11.22VVRR224 pKa = 11.84QWTFMLLGDD233 pKa = 5.01ADD235 pKa = 3.86ACATGSEE242 pKa = 4.14AFAIDD247 pKa = 4.36EE248 pKa = 4.22YY249 pKa = 11.92VKK251 pKa = 11.14LNLPTCWLKK260 pKa = 11.49ANTDD264 pKa = 3.09GSFNSMEE271 pKa = 4.12KK272 pKa = 10.72GGLCLYY278 pKa = 9.72VQGSSALIPVLFPVIRR294 pKa = 11.84LYY296 pKa = 11.09FEE298 pKa = 4.54DD299 pKa = 3.5QQ300 pKa = 3.08
MM1 pKa = 7.79PEE3 pKa = 4.12KK4 pKa = 10.39FLADD8 pKa = 4.74GISFHH13 pKa = 5.83HH14 pKa = 6.59HH15 pKa = 6.65KK16 pKa = 10.71KK17 pKa = 10.35LLCCAYY23 pKa = 9.68ILGINRR29 pKa = 11.84AKK31 pKa = 10.64GLRR34 pKa = 11.84IMSMYY39 pKa = 9.75YY40 pKa = 9.85RR41 pKa = 11.84KK42 pKa = 10.22KK43 pKa = 8.89KK44 pKa = 8.71TGGGGAARR52 pKa = 11.84GSVKK56 pKa = 10.42GYY58 pKa = 9.84AAAATRR64 pKa = 11.84VPGTAGKK71 pKa = 10.38AKK73 pKa = 10.26LAAARR78 pKa = 11.84NASKK82 pKa = 10.18SAQMARR88 pKa = 11.84YY89 pKa = 8.52VGRR92 pKa = 11.84RR93 pKa = 11.84SNVPGKK99 pKa = 10.08DD100 pKa = 2.7NHH102 pKa = 6.33TFDD105 pKa = 4.72PVVLNTNYY113 pKa = 10.07LQCDD117 pKa = 3.3NATAVAVSASGYY129 pKa = 6.97ITPGSSAIVLNQVSQGTSSTQRR151 pKa = 11.84IGRR154 pKa = 11.84KK155 pKa = 8.87ILMTAIHH162 pKa = 5.96IRR164 pKa = 11.84AYY166 pKa = 10.67LRR168 pKa = 11.84NTTASADD175 pKa = 3.61LNMVRR180 pKa = 11.84VALVYY185 pKa = 10.18IPEE188 pKa = 4.18MDD190 pKa = 3.25RR191 pKa = 11.84TTTTMPPQNAIWTSQDD207 pKa = 2.72ARR209 pKa = 11.84EE210 pKa = 4.5LRR212 pKa = 11.84VIDD215 pKa = 3.37NADD218 pKa = 3.49RR219 pKa = 11.84FKK221 pKa = 11.22VVRR224 pKa = 11.84QWTFMLLGDD233 pKa = 5.01ADD235 pKa = 3.86ACATGSEE242 pKa = 4.14AFAIDD247 pKa = 4.36EE248 pKa = 4.22YY249 pKa = 11.92VKK251 pKa = 11.14LNLPTCWLKK260 pKa = 11.49ANTDD264 pKa = 3.09GSFNSMEE271 pKa = 4.12KK272 pKa = 10.72GGLCLYY278 pKa = 9.72VQGSSALIPVLFPVIRR294 pKa = 11.84LYY296 pKa = 11.09FEE298 pKa = 4.54DD299 pKa = 3.5QQ300 pKa = 3.08
Molecular weight: 32.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
628 |
300 |
328 |
314.0 |
35.14 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.032 ± 1.511 | 1.752 ± 0.163 |
5.255 ± 0.386 | 4.618 ± 1.5 |
3.662 ± 0.435 | 7.803 ± 0.089 |
2.07 ± 0.265 | 4.299 ± 0.679 |
6.051 ± 0.252 | 7.803 ± 0.13 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.866 ± 0.307 | 3.822 ± 0.774 |
5.255 ± 1.043 | 3.503 ± 0.112 |
6.847 ± 0.118 | 5.414 ± 0.822 |
5.414 ± 1.041 | 6.847 ± 0.118 |
2.07 ± 0.703 | 4.618 ± 0.406 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
