
Peptoclostridium acidaminophilum DSM 3953
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Peptostreptococcaceae; Peptoclostridium; Peptoclostridium acidaminophilum
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
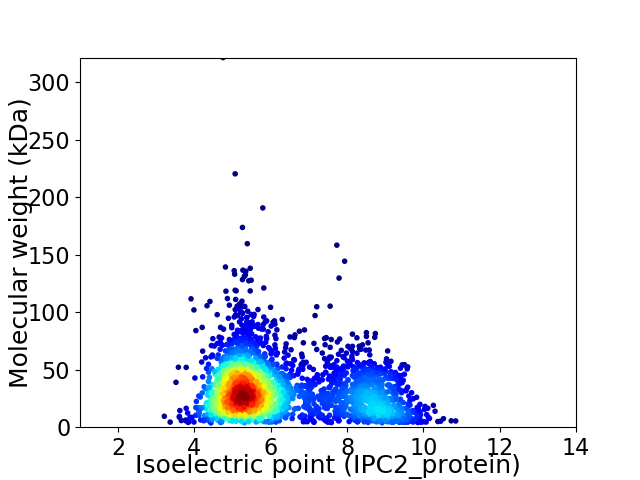
Virtual 2D-PAGE plot for 2843 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W8T706|W8T706_PEPAC Elongation factor Ts OS=Peptoclostridium acidaminophilum DSM 3953 OX=1286171 GN=tsf PE=3 SV=1
MM1 pKa = 7.35IAKK4 pKa = 9.76KK5 pKa = 10.06AARR8 pKa = 11.84YY9 pKa = 9.02LLAITICIVCFGSSIFASSNDD30 pKa = 3.35NPNAEE35 pKa = 4.43KK36 pKa = 10.82LKK38 pKa = 10.73ALKK41 pKa = 8.64KK42 pKa = 8.13TQATEE47 pKa = 4.03EE48 pKa = 4.33KK49 pKa = 10.29LGEE52 pKa = 4.33YY53 pKa = 10.14YY54 pKa = 10.77DD55 pKa = 4.3SLDD58 pKa = 3.38TGSAQDD64 pKa = 3.27IAVGVSTEE72 pKa = 4.64DD73 pKa = 3.66IIAAYY78 pKa = 10.51SNSTYY83 pKa = 10.35RR84 pKa = 11.84INIQEE89 pKa = 4.1SGFYY93 pKa = 10.44HH94 pKa = 7.04IDD96 pKa = 3.31LGLTEE101 pKa = 5.17DD102 pKa = 3.96ADD104 pKa = 4.44LFVEE108 pKa = 5.07TYY110 pKa = 10.54DD111 pKa = 3.81EE112 pKa = 4.14SLEE115 pKa = 4.06YY116 pKa = 10.74VDD118 pKa = 4.23WGSMVYY124 pKa = 10.4LQGNADD130 pKa = 3.62YY131 pKa = 11.02YY132 pKa = 10.52IYY134 pKa = 10.66LDD136 pKa = 4.38NYY138 pKa = 9.73YY139 pKa = 10.52QDD141 pKa = 3.67TDD143 pKa = 3.79TQVNICVQKK152 pKa = 10.33VQPTMYY158 pKa = 10.18EE159 pKa = 3.84LGSDD163 pKa = 4.13LVADD167 pKa = 4.69GVMQDD172 pKa = 4.5GEE174 pKa = 4.17VHH176 pKa = 5.81MYY178 pKa = 9.04EE179 pKa = 4.25FSVGEE184 pKa = 3.78EE185 pKa = 3.93ALYY188 pKa = 10.36EE189 pKa = 3.98IYY191 pKa = 10.87GCDD194 pKa = 5.34FILLDD199 pKa = 3.84EE200 pKa = 4.47NFNRR204 pKa = 11.84IEE206 pKa = 4.1CDD208 pKa = 3.69YY209 pKa = 10.35EE210 pKa = 4.93CSLSPDD216 pKa = 3.78EE217 pKa = 4.75IYY219 pKa = 11.13YY220 pKa = 10.21IAVADD225 pKa = 3.79KK226 pKa = 10.67GYY228 pKa = 10.39SAEE231 pKa = 4.23EE232 pKa = 3.73YY233 pKa = 8.42WYY235 pKa = 10.34DD236 pKa = 3.84VIVEE240 pKa = 4.74NVDD243 pKa = 2.67DD244 pKa = 4.97WYY246 pKa = 10.68DD247 pKa = 3.51DD248 pKa = 3.87DD249 pKa = 5.84YY250 pKa = 11.42EE251 pKa = 4.36YY252 pKa = 11.32TDD254 pKa = 5.22VEE256 pKa = 4.37PQGNDD261 pKa = 3.32YY262 pKa = 10.97IEE264 pKa = 4.42YY265 pKa = 10.59AVNEE269 pKa = 4.04YY270 pKa = 10.49DD271 pKa = 3.8GKK273 pKa = 9.7FTIGTRR279 pKa = 11.84EE280 pKa = 4.2GTSNPNDD287 pKa = 3.98DD288 pKa = 4.12NKK290 pKa = 11.35KK291 pKa = 8.95MLFGHH296 pKa = 7.5PYY298 pKa = 9.34PGTSFTTINLDD309 pKa = 3.33GEE311 pKa = 4.6YY312 pKa = 10.97AKK314 pKa = 9.28YY315 pKa = 9.06TPDD318 pKa = 3.65AEE320 pKa = 4.52SSISSGTTNMSEE332 pKa = 3.77QTMGDD337 pKa = 3.36VSVKK341 pKa = 10.07QVLSIVKK348 pKa = 10.16NSATQKK354 pKa = 10.36EE355 pKa = 4.51DD356 pKa = 3.22VVQIKK361 pKa = 10.46YY362 pKa = 10.26IVTNNDD368 pKa = 3.11GASHH372 pKa = 6.8DD373 pKa = 3.75VGVRR377 pKa = 11.84IMMDD381 pKa = 3.11TMLGDD386 pKa = 3.67NDD388 pKa = 4.0SAPFKK393 pKa = 10.8VPNVGNVLYY402 pKa = 10.32EE403 pKa = 4.03RR404 pKa = 11.84EE405 pKa = 4.13YY406 pKa = 10.5TGADD410 pKa = 3.29VPVYY414 pKa = 8.9WQAFDD419 pKa = 4.96SIEE422 pKa = 4.08NPTIVSQGTLRR433 pKa = 11.84DD434 pKa = 4.2GYY436 pKa = 9.21NTAPDD441 pKa = 3.76KK442 pKa = 11.4VQFASWGDD450 pKa = 3.36ITDD453 pKa = 3.88TLWDD457 pKa = 4.0YY458 pKa = 10.86EE459 pKa = 4.17VSEE462 pKa = 4.18YY463 pKa = 10.93QEE465 pKa = 4.63IGDD468 pKa = 4.22SSVAVYY474 pKa = 8.74WNPITIGPGARR485 pKa = 11.84KK486 pKa = 10.19DD487 pKa = 3.46FVTYY491 pKa = 10.49YY492 pKa = 10.89GLSEE496 pKa = 4.47FSANAASGMVLGVTAPSEE514 pKa = 3.94LVLDD518 pKa = 4.1KK519 pKa = 11.42EE520 pKa = 5.05EE521 pKa = 4.04YY522 pKa = 10.07TPDD525 pKa = 3.63PFIVTAYY532 pKa = 9.13IVNSGEE538 pKa = 3.9EE539 pKa = 4.0TSANTVVKK547 pKa = 10.55INLPEE552 pKa = 4.15GLNLYY557 pKa = 10.13SGSSEE562 pKa = 4.24VNVGDD567 pKa = 3.89VLPGEE572 pKa = 4.42EE573 pKa = 5.37KK574 pKa = 10.63LVSWYY579 pKa = 8.13VTAVKK584 pKa = 10.5RR585 pKa = 11.84EE586 pKa = 4.18TEE588 pKa = 3.87DD589 pKa = 3.11TLAYY593 pKa = 10.18SVEE596 pKa = 4.02LSSDD600 pKa = 3.22GVAGAFEE607 pKa = 4.39QKK609 pKa = 10.39HH610 pKa = 4.32ITVPSVSAIKK620 pKa = 10.14FADD623 pKa = 4.02PGLEE627 pKa = 3.85QAIRR631 pKa = 11.84DD632 pKa = 3.77ALGQSEE638 pKa = 4.33GAITKK643 pKa = 9.54TDD645 pKa = 3.83LEE647 pKa = 5.02SITALNAEE655 pKa = 4.3NLEE658 pKa = 3.99ISDD661 pKa = 4.1LSGIEE666 pKa = 3.78YY667 pKa = 10.44LVNLEE672 pKa = 4.0EE673 pKa = 6.02LKK675 pKa = 11.15LNYY678 pKa = 10.47NNIDD682 pKa = 3.85DD683 pKa = 4.97LNLVVDD689 pKa = 4.03NLQNLEE695 pKa = 3.97RR696 pKa = 11.84LEE698 pKa = 4.06LRR700 pKa = 11.84GNNIEE705 pKa = 4.52NIFISFFMQNSVVLAQEE722 pKa = 4.05RR723 pKa = 11.84LGATLGKK730 pKa = 10.19LPNLSYY736 pKa = 11.54LDD738 pKa = 3.89LSEE741 pKa = 5.38NNINNIEE748 pKa = 4.3GIEE751 pKa = 4.06NFTNIEE757 pKa = 4.12TLNLSANKK765 pKa = 9.65IDD767 pKa = 6.04DD768 pKa = 3.81IANLAGLSGMKK779 pKa = 9.98SLNISGNQVEE789 pKa = 5.66DD790 pKa = 3.32ITPLLEE796 pKa = 4.21VKK798 pKa = 9.47TLEE801 pKa = 4.64SVKK804 pKa = 10.85LQDD807 pKa = 3.7NPVVEE812 pKa = 4.51IPSEE816 pKa = 3.61ISEE819 pKa = 4.37IINVKK824 pKa = 10.47SLGKK828 pKa = 8.8KK829 pKa = 8.5TGITSNKK836 pKa = 5.68VWRR839 pKa = 11.84VQFSKK844 pKa = 11.13GLDD847 pKa = 3.09EE848 pKa = 4.04MLNYY852 pKa = 10.45NNYY855 pKa = 7.89VTVKK859 pKa = 10.39DD860 pKa = 3.66SLGNIVDD867 pKa = 3.57VDD869 pKa = 3.57VTYY872 pKa = 11.27SKK874 pKa = 11.3NEE876 pKa = 3.63KK877 pKa = 10.29ALLVTPKK884 pKa = 10.17SSYY887 pKa = 10.7SAGEE891 pKa = 4.13TYY893 pKa = 10.53SIVIGEE899 pKa = 4.89GILSEE904 pKa = 4.26KK905 pKa = 11.13GEE907 pKa = 4.37TLPEE911 pKa = 3.88AVRR914 pKa = 11.84MVFTVAQQ921 pKa = 3.65
MM1 pKa = 7.35IAKK4 pKa = 9.76KK5 pKa = 10.06AARR8 pKa = 11.84YY9 pKa = 9.02LLAITICIVCFGSSIFASSNDD30 pKa = 3.35NPNAEE35 pKa = 4.43KK36 pKa = 10.82LKK38 pKa = 10.73ALKK41 pKa = 8.64KK42 pKa = 8.13TQATEE47 pKa = 4.03EE48 pKa = 4.33KK49 pKa = 10.29LGEE52 pKa = 4.33YY53 pKa = 10.14YY54 pKa = 10.77DD55 pKa = 4.3SLDD58 pKa = 3.38TGSAQDD64 pKa = 3.27IAVGVSTEE72 pKa = 4.64DD73 pKa = 3.66IIAAYY78 pKa = 10.51SNSTYY83 pKa = 10.35RR84 pKa = 11.84INIQEE89 pKa = 4.1SGFYY93 pKa = 10.44HH94 pKa = 7.04IDD96 pKa = 3.31LGLTEE101 pKa = 5.17DD102 pKa = 3.96ADD104 pKa = 4.44LFVEE108 pKa = 5.07TYY110 pKa = 10.54DD111 pKa = 3.81EE112 pKa = 4.14SLEE115 pKa = 4.06YY116 pKa = 10.74VDD118 pKa = 4.23WGSMVYY124 pKa = 10.4LQGNADD130 pKa = 3.62YY131 pKa = 11.02YY132 pKa = 10.52IYY134 pKa = 10.66LDD136 pKa = 4.38NYY138 pKa = 9.73YY139 pKa = 10.52QDD141 pKa = 3.67TDD143 pKa = 3.79TQVNICVQKK152 pKa = 10.33VQPTMYY158 pKa = 10.18EE159 pKa = 3.84LGSDD163 pKa = 4.13LVADD167 pKa = 4.69GVMQDD172 pKa = 4.5GEE174 pKa = 4.17VHH176 pKa = 5.81MYY178 pKa = 9.04EE179 pKa = 4.25FSVGEE184 pKa = 3.78EE185 pKa = 3.93ALYY188 pKa = 10.36EE189 pKa = 3.98IYY191 pKa = 10.87GCDD194 pKa = 5.34FILLDD199 pKa = 3.84EE200 pKa = 4.47NFNRR204 pKa = 11.84IEE206 pKa = 4.1CDD208 pKa = 3.69YY209 pKa = 10.35EE210 pKa = 4.93CSLSPDD216 pKa = 3.78EE217 pKa = 4.75IYY219 pKa = 11.13YY220 pKa = 10.21IAVADD225 pKa = 3.79KK226 pKa = 10.67GYY228 pKa = 10.39SAEE231 pKa = 4.23EE232 pKa = 3.73YY233 pKa = 8.42WYY235 pKa = 10.34DD236 pKa = 3.84VIVEE240 pKa = 4.74NVDD243 pKa = 2.67DD244 pKa = 4.97WYY246 pKa = 10.68DD247 pKa = 3.51DD248 pKa = 3.87DD249 pKa = 5.84YY250 pKa = 11.42EE251 pKa = 4.36YY252 pKa = 11.32TDD254 pKa = 5.22VEE256 pKa = 4.37PQGNDD261 pKa = 3.32YY262 pKa = 10.97IEE264 pKa = 4.42YY265 pKa = 10.59AVNEE269 pKa = 4.04YY270 pKa = 10.49DD271 pKa = 3.8GKK273 pKa = 9.7FTIGTRR279 pKa = 11.84EE280 pKa = 4.2GTSNPNDD287 pKa = 3.98DD288 pKa = 4.12NKK290 pKa = 11.35KK291 pKa = 8.95MLFGHH296 pKa = 7.5PYY298 pKa = 9.34PGTSFTTINLDD309 pKa = 3.33GEE311 pKa = 4.6YY312 pKa = 10.97AKK314 pKa = 9.28YY315 pKa = 9.06TPDD318 pKa = 3.65AEE320 pKa = 4.52SSISSGTTNMSEE332 pKa = 3.77QTMGDD337 pKa = 3.36VSVKK341 pKa = 10.07QVLSIVKK348 pKa = 10.16NSATQKK354 pKa = 10.36EE355 pKa = 4.51DD356 pKa = 3.22VVQIKK361 pKa = 10.46YY362 pKa = 10.26IVTNNDD368 pKa = 3.11GASHH372 pKa = 6.8DD373 pKa = 3.75VGVRR377 pKa = 11.84IMMDD381 pKa = 3.11TMLGDD386 pKa = 3.67NDD388 pKa = 4.0SAPFKK393 pKa = 10.8VPNVGNVLYY402 pKa = 10.32EE403 pKa = 4.03RR404 pKa = 11.84EE405 pKa = 4.13YY406 pKa = 10.5TGADD410 pKa = 3.29VPVYY414 pKa = 8.9WQAFDD419 pKa = 4.96SIEE422 pKa = 4.08NPTIVSQGTLRR433 pKa = 11.84DD434 pKa = 4.2GYY436 pKa = 9.21NTAPDD441 pKa = 3.76KK442 pKa = 11.4VQFASWGDD450 pKa = 3.36ITDD453 pKa = 3.88TLWDD457 pKa = 4.0YY458 pKa = 10.86EE459 pKa = 4.17VSEE462 pKa = 4.18YY463 pKa = 10.93QEE465 pKa = 4.63IGDD468 pKa = 4.22SSVAVYY474 pKa = 8.74WNPITIGPGARR485 pKa = 11.84KK486 pKa = 10.19DD487 pKa = 3.46FVTYY491 pKa = 10.49YY492 pKa = 10.89GLSEE496 pKa = 4.47FSANAASGMVLGVTAPSEE514 pKa = 3.94LVLDD518 pKa = 4.1KK519 pKa = 11.42EE520 pKa = 5.05EE521 pKa = 4.04YY522 pKa = 10.07TPDD525 pKa = 3.63PFIVTAYY532 pKa = 9.13IVNSGEE538 pKa = 3.9EE539 pKa = 4.0TSANTVVKK547 pKa = 10.55INLPEE552 pKa = 4.15GLNLYY557 pKa = 10.13SGSSEE562 pKa = 4.24VNVGDD567 pKa = 3.89VLPGEE572 pKa = 4.42EE573 pKa = 5.37KK574 pKa = 10.63LVSWYY579 pKa = 8.13VTAVKK584 pKa = 10.5RR585 pKa = 11.84EE586 pKa = 4.18TEE588 pKa = 3.87DD589 pKa = 3.11TLAYY593 pKa = 10.18SVEE596 pKa = 4.02LSSDD600 pKa = 3.22GVAGAFEE607 pKa = 4.39QKK609 pKa = 10.39HH610 pKa = 4.32ITVPSVSAIKK620 pKa = 10.14FADD623 pKa = 4.02PGLEE627 pKa = 3.85QAIRR631 pKa = 11.84DD632 pKa = 3.77ALGQSEE638 pKa = 4.33GAITKK643 pKa = 9.54TDD645 pKa = 3.83LEE647 pKa = 5.02SITALNAEE655 pKa = 4.3NLEE658 pKa = 3.99ISDD661 pKa = 4.1LSGIEE666 pKa = 3.78YY667 pKa = 10.44LVNLEE672 pKa = 4.0EE673 pKa = 6.02LKK675 pKa = 11.15LNYY678 pKa = 10.47NNIDD682 pKa = 3.85DD683 pKa = 4.97LNLVVDD689 pKa = 4.03NLQNLEE695 pKa = 3.97RR696 pKa = 11.84LEE698 pKa = 4.06LRR700 pKa = 11.84GNNIEE705 pKa = 4.52NIFISFFMQNSVVLAQEE722 pKa = 4.05RR723 pKa = 11.84LGATLGKK730 pKa = 10.19LPNLSYY736 pKa = 11.54LDD738 pKa = 3.89LSEE741 pKa = 5.38NNINNIEE748 pKa = 4.3GIEE751 pKa = 4.06NFTNIEE757 pKa = 4.12TLNLSANKK765 pKa = 9.65IDD767 pKa = 6.04DD768 pKa = 3.81IANLAGLSGMKK779 pKa = 9.98SLNISGNQVEE789 pKa = 5.66DD790 pKa = 3.32ITPLLEE796 pKa = 4.21VKK798 pKa = 9.47TLEE801 pKa = 4.64SVKK804 pKa = 10.85LQDD807 pKa = 3.7NPVVEE812 pKa = 4.51IPSEE816 pKa = 3.61ISEE819 pKa = 4.37IINVKK824 pKa = 10.47SLGKK828 pKa = 8.8KK829 pKa = 8.5TGITSNKK836 pKa = 5.68VWRR839 pKa = 11.84VQFSKK844 pKa = 11.13GLDD847 pKa = 3.09EE848 pKa = 4.04MLNYY852 pKa = 10.45NNYY855 pKa = 7.89VTVKK859 pKa = 10.39DD860 pKa = 3.66SLGNIVDD867 pKa = 3.57VDD869 pKa = 3.57VTYY872 pKa = 11.27SKK874 pKa = 11.3NEE876 pKa = 3.63KK877 pKa = 10.29ALLVTPKK884 pKa = 10.17SSYY887 pKa = 10.7SAGEE891 pKa = 4.13TYY893 pKa = 10.53SIVIGEE899 pKa = 4.89GILSEE904 pKa = 4.26KK905 pKa = 11.13GEE907 pKa = 4.37TLPEE911 pKa = 3.88AVRR914 pKa = 11.84MVFTVAQQ921 pKa = 3.65
Molecular weight: 101.92 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W8TK11|W8TK11_PEPAC Pyruvate carboxylase OS=Peptoclostridium acidaminophilum DSM 3953 OX=1286171 GN=pyc PE=4 SV=1
MM1 pKa = 7.04IRR3 pKa = 11.84WYY5 pKa = 9.68ATGTSIAGRR14 pKa = 11.84RR15 pKa = 11.84FAIFILLTPSMLRR28 pKa = 11.84PIPNMSKK35 pKa = 10.82LPTTLISDD43 pKa = 4.08ITCCVSMPLRR53 pKa = 11.84NPAPRR58 pKa = 11.84VIAPWYY64 pKa = 9.63ISTEE68 pKa = 3.93MADD71 pKa = 4.32IITPMPIVDD80 pKa = 4.29ANIMADD86 pKa = 3.62TPSRR90 pKa = 11.84TDD92 pKa = 3.45FANSVLWSPLSPLSSAPTIAIAPTQYY118 pKa = 10.84ISVQLTKK125 pKa = 10.41PSATLPPLPLKK136 pKa = 10.58SSSRR140 pKa = 11.84RR141 pKa = 11.84SPNLISLWSISITSPITPPMTMEE164 pKa = 5.23NITSSRR170 pKa = 11.84LASSRR175 pKa = 11.84APLIPMRR182 pKa = 11.84TTHH185 pKa = 6.13SAIPFITVSLNFSVFIPLIIIPAKK209 pKa = 10.58LPATIAATLTSVPIIQSPPFF229 pKa = 3.97
MM1 pKa = 7.04IRR3 pKa = 11.84WYY5 pKa = 9.68ATGTSIAGRR14 pKa = 11.84RR15 pKa = 11.84FAIFILLTPSMLRR28 pKa = 11.84PIPNMSKK35 pKa = 10.82LPTTLISDD43 pKa = 4.08ITCCVSMPLRR53 pKa = 11.84NPAPRR58 pKa = 11.84VIAPWYY64 pKa = 9.63ISTEE68 pKa = 3.93MADD71 pKa = 4.32IITPMPIVDD80 pKa = 4.29ANIMADD86 pKa = 3.62TPSRR90 pKa = 11.84TDD92 pKa = 3.45FANSVLWSPLSPLSSAPTIAIAPTQYY118 pKa = 10.84ISVQLTKK125 pKa = 10.41PSATLPPLPLKK136 pKa = 10.58SSSRR140 pKa = 11.84RR141 pKa = 11.84SPNLISLWSISITSPITPPMTMEE164 pKa = 5.23NITSSRR170 pKa = 11.84LASSRR175 pKa = 11.84APLIPMRR182 pKa = 11.84TTHH185 pKa = 6.13SAIPFITVSLNFSVFIPLIIIPAKK209 pKa = 10.58LPATIAATLTSVPIIQSPPFF229 pKa = 3.97
Molecular weight: 24.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
861762 |
35 |
2802 |
303.1 |
33.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.559 ± 0.057 | 1.18 ± 0.021 |
5.644 ± 0.044 | 7.775 ± 0.059 |
4.055 ± 0.034 | 7.336 ± 0.043 |
1.592 ± 0.016 | 8.262 ± 0.044 |
7.456 ± 0.042 | 8.854 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.094 ± 0.021 | 4.271 ± 0.029 |
3.118 ± 0.024 | 2.708 ± 0.022 |
4.527 ± 0.035 | 6.778 ± 0.042 |
4.572 ± 0.028 | 6.903 ± 0.04 |
0.732 ± 0.015 | 3.584 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
