
Streptococcus phage Javan68
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.21
Get precalculated fractions of proteins
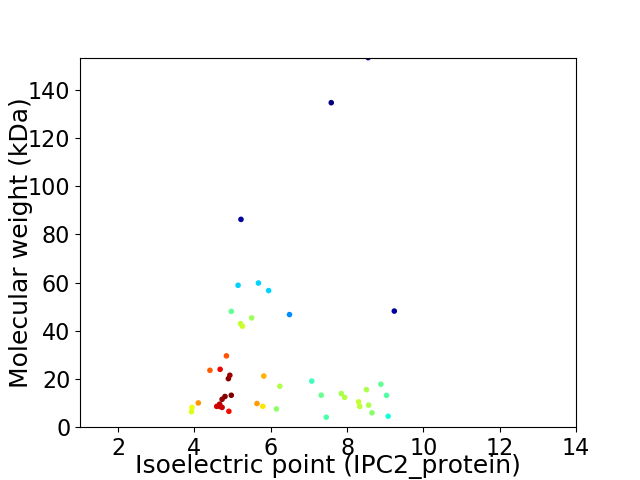
Virtual 2D-PAGE plot for 46 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
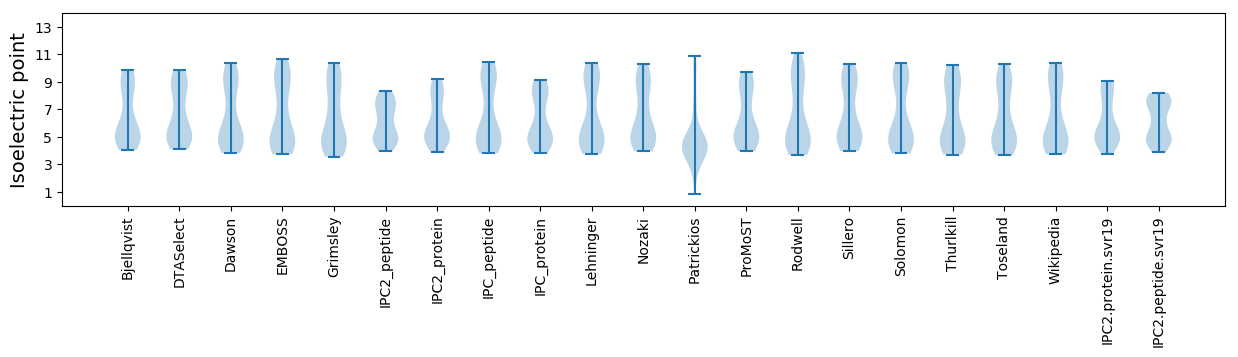
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4D6BD53|A0A4D6BD53_9CAUD Tail assembly protein OS=Streptococcus phage Javan68 OX=2548301 GN=Javan68_0005 PE=4 SV=1
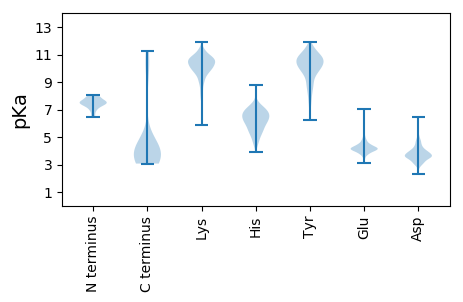
MM1 pKa = 7.47HH2 pKa = 7.02GKK4 pKa = 9.97KK5 pKa = 10.18YY6 pKa = 11.22YY7 pKa = 10.87EE8 pKa = 4.36MDD10 pKa = 4.06DD11 pKa = 4.32EE12 pKa = 5.73INDD15 pKa = 3.6VPHH18 pKa = 7.01FGLTYY23 pKa = 10.58SEE25 pKa = 5.04FEE27 pKa = 4.04GDD29 pKa = 3.09DD30 pKa = 3.28WVIGFDD36 pKa = 4.01CNHH39 pKa = 6.74AFDD42 pKa = 4.46TPATNTVEE50 pKa = 4.26FVEE53 pKa = 4.92GNIKK57 pKa = 10.43EE58 pKa = 4.22IVDD61 pKa = 4.77TILEE65 pKa = 4.3IYY67 pKa = 10.22PEE69 pKa = 4.71GEE71 pKa = 3.79
MM1 pKa = 7.47HH2 pKa = 7.02GKK4 pKa = 9.97KK5 pKa = 10.18YY6 pKa = 11.22YY7 pKa = 10.87EE8 pKa = 4.36MDD10 pKa = 4.06DD11 pKa = 4.32EE12 pKa = 5.73INDD15 pKa = 3.6VPHH18 pKa = 7.01FGLTYY23 pKa = 10.58SEE25 pKa = 5.04FEE27 pKa = 4.04GDD29 pKa = 3.09DD30 pKa = 3.28WVIGFDD36 pKa = 4.01CNHH39 pKa = 6.74AFDD42 pKa = 4.46TPATNTVEE50 pKa = 4.26FVEE53 pKa = 4.92GNIKK57 pKa = 10.43EE58 pKa = 4.22IVDD61 pKa = 4.77TILEE65 pKa = 4.3IYY67 pKa = 10.22PEE69 pKa = 4.71GEE71 pKa = 3.79
Molecular weight: 8.19 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D6BHF2|A0A4D6BHF2_9CAUD Tail length tape-measure protein OS=Streptococcus phage Javan68 OX=2548301 GN=Javan68_0007 PE=4 SV=1
MM1 pKa = 7.92PISFLKK7 pKa = 10.46RR8 pKa = 11.84SCPEE12 pKa = 3.38VAFIEE17 pKa = 4.26RR18 pKa = 11.84KK19 pKa = 8.7KK20 pKa = 10.96IKK22 pKa = 9.46MKK24 pKa = 8.97YY25 pKa = 8.59TKK27 pKa = 9.33TKK29 pKa = 9.21YY30 pKa = 9.68PNIYY34 pKa = 9.56TYY36 pKa = 8.37EE37 pKa = 4.05TTKK40 pKa = 10.58GKK42 pKa = 10.05RR43 pKa = 11.84YY44 pKa = 7.98YY45 pKa = 10.47VRR47 pKa = 11.84RR48 pKa = 11.84SFMFQGKK55 pKa = 8.49KK56 pKa = 9.49QEE58 pKa = 4.29ANKK61 pKa = 10.58SSLKK65 pKa = 9.91TISEE69 pKa = 4.01ARR71 pKa = 11.84AEE73 pKa = 3.85LAKK76 pKa = 10.16IEE78 pKa = 4.76KK79 pKa = 10.07RR80 pKa = 11.84IADD83 pKa = 3.43QTIAINPNVTVEE95 pKa = 4.29EE96 pKa = 4.36YY97 pKa = 9.87WEE99 pKa = 3.94KK100 pKa = 10.55FYY102 pKa = 11.24EE103 pKa = 4.17KK104 pKa = 10.03RR105 pKa = 11.84VEE107 pKa = 4.14TGRR110 pKa = 11.84WTPNTEE116 pKa = 3.72VYY118 pKa = 10.56YY119 pKa = 10.94KK120 pKa = 9.96STFKK124 pKa = 11.07LHH126 pKa = 7.08ILPFWGKK133 pKa = 8.05TKK135 pKa = 10.8LKK137 pKa = 9.7NLSRR141 pKa = 11.84NEE143 pKa = 4.09YY144 pKa = 8.91EE145 pKa = 3.86KK146 pKa = 10.96HH147 pKa = 5.37IANLLKK153 pKa = 10.4VRR155 pKa = 11.84PKK157 pKa = 9.53ASVRR161 pKa = 11.84IIHH164 pKa = 5.87SCFMTMLNDD173 pKa = 4.9AIMNGNISANRR184 pKa = 11.84LNGIYY189 pKa = 10.61VGDD192 pKa = 3.78SLIAKK197 pKa = 8.69KK198 pKa = 10.43NKK200 pKa = 10.08RR201 pKa = 11.84ITLDD205 pKa = 3.15QFQIWMQEE213 pKa = 3.72AEE215 pKa = 4.24KK216 pKa = 11.34VMDD219 pKa = 3.38KK220 pKa = 10.8TFFGLTYY227 pKa = 10.56LAIFGLRR234 pKa = 11.84RR235 pKa = 11.84GEE237 pKa = 3.9ILGLRR242 pKa = 11.84HH243 pKa = 6.65CDD245 pKa = 3.22VNINVNGRR253 pKa = 11.84AVLHH257 pKa = 6.04LQDD260 pKa = 4.48SRR262 pKa = 11.84GNHH265 pKa = 5.01TKK267 pKa = 10.55NGRR270 pKa = 11.84HH271 pKa = 4.77GLKK274 pKa = 9.47TKK276 pKa = 10.63SSEE279 pKa = 3.84RR280 pKa = 11.84WVTLDD285 pKa = 3.23NTGTEE290 pKa = 4.28LLLSLMSNAEE300 pKa = 4.01AVKK303 pKa = 10.59KK304 pKa = 9.99KK305 pKa = 10.82LNIIKK310 pKa = 9.89EE311 pKa = 4.24VEE313 pKa = 3.46WDD315 pKa = 4.07YY316 pKa = 11.84ISIDD320 pKa = 3.38QKK322 pKa = 11.51GRR324 pKa = 11.84LINPNKK330 pKa = 10.15LNRR333 pKa = 11.84EE334 pKa = 4.08FEE336 pKa = 4.24RR337 pKa = 11.84VNDD340 pKa = 3.58VVGFRR345 pKa = 11.84VTPHH349 pKa = 5.79MLRR352 pKa = 11.84HH353 pKa = 6.02FFTTQSIIAGVPMEE367 pKa = 5.08ALSQALGHH375 pKa = 6.09TKK377 pKa = 9.66MYY379 pKa = 7.84MTDD382 pKa = 3.49KK383 pKa = 10.69YY384 pKa = 11.15NQVHH388 pKa = 7.18DD389 pKa = 4.28EE390 pKa = 4.27LSAQVSDD397 pKa = 4.41AFIAQISPRR406 pKa = 11.84NSPTNLHH413 pKa = 6.35ILL415 pKa = 3.73
MM1 pKa = 7.92PISFLKK7 pKa = 10.46RR8 pKa = 11.84SCPEE12 pKa = 3.38VAFIEE17 pKa = 4.26RR18 pKa = 11.84KK19 pKa = 8.7KK20 pKa = 10.96IKK22 pKa = 9.46MKK24 pKa = 8.97YY25 pKa = 8.59TKK27 pKa = 9.33TKK29 pKa = 9.21YY30 pKa = 9.68PNIYY34 pKa = 9.56TYY36 pKa = 8.37EE37 pKa = 4.05TTKK40 pKa = 10.58GKK42 pKa = 10.05RR43 pKa = 11.84YY44 pKa = 7.98YY45 pKa = 10.47VRR47 pKa = 11.84RR48 pKa = 11.84SFMFQGKK55 pKa = 8.49KK56 pKa = 9.49QEE58 pKa = 4.29ANKK61 pKa = 10.58SSLKK65 pKa = 9.91TISEE69 pKa = 4.01ARR71 pKa = 11.84AEE73 pKa = 3.85LAKK76 pKa = 10.16IEE78 pKa = 4.76KK79 pKa = 10.07RR80 pKa = 11.84IADD83 pKa = 3.43QTIAINPNVTVEE95 pKa = 4.29EE96 pKa = 4.36YY97 pKa = 9.87WEE99 pKa = 3.94KK100 pKa = 10.55FYY102 pKa = 11.24EE103 pKa = 4.17KK104 pKa = 10.03RR105 pKa = 11.84VEE107 pKa = 4.14TGRR110 pKa = 11.84WTPNTEE116 pKa = 3.72VYY118 pKa = 10.56YY119 pKa = 10.94KK120 pKa = 9.96STFKK124 pKa = 11.07LHH126 pKa = 7.08ILPFWGKK133 pKa = 8.05TKK135 pKa = 10.8LKK137 pKa = 9.7NLSRR141 pKa = 11.84NEE143 pKa = 4.09YY144 pKa = 8.91EE145 pKa = 3.86KK146 pKa = 10.96HH147 pKa = 5.37IANLLKK153 pKa = 10.4VRR155 pKa = 11.84PKK157 pKa = 9.53ASVRR161 pKa = 11.84IIHH164 pKa = 5.87SCFMTMLNDD173 pKa = 4.9AIMNGNISANRR184 pKa = 11.84LNGIYY189 pKa = 10.61VGDD192 pKa = 3.78SLIAKK197 pKa = 8.69KK198 pKa = 10.43NKK200 pKa = 10.08RR201 pKa = 11.84ITLDD205 pKa = 3.15QFQIWMQEE213 pKa = 3.72AEE215 pKa = 4.24KK216 pKa = 11.34VMDD219 pKa = 3.38KK220 pKa = 10.8TFFGLTYY227 pKa = 10.56LAIFGLRR234 pKa = 11.84RR235 pKa = 11.84GEE237 pKa = 3.9ILGLRR242 pKa = 11.84HH243 pKa = 6.65CDD245 pKa = 3.22VNINVNGRR253 pKa = 11.84AVLHH257 pKa = 6.04LQDD260 pKa = 4.48SRR262 pKa = 11.84GNHH265 pKa = 5.01TKK267 pKa = 10.55NGRR270 pKa = 11.84HH271 pKa = 4.77GLKK274 pKa = 9.47TKK276 pKa = 10.63SSEE279 pKa = 3.84RR280 pKa = 11.84WVTLDD285 pKa = 3.23NTGTEE290 pKa = 4.28LLLSLMSNAEE300 pKa = 4.01AVKK303 pKa = 10.59KK304 pKa = 9.99KK305 pKa = 10.82LNIIKK310 pKa = 9.89EE311 pKa = 4.24VEE313 pKa = 3.46WDD315 pKa = 4.07YY316 pKa = 11.84ISIDD320 pKa = 3.38QKK322 pKa = 11.51GRR324 pKa = 11.84LINPNKK330 pKa = 10.15LNRR333 pKa = 11.84EE334 pKa = 4.08FEE336 pKa = 4.24RR337 pKa = 11.84VNDD340 pKa = 3.58VVGFRR345 pKa = 11.84VTPHH349 pKa = 5.79MLRR352 pKa = 11.84HH353 pKa = 6.02FFTTQSIIAGVPMEE367 pKa = 5.08ALSQALGHH375 pKa = 6.09TKK377 pKa = 9.66MYY379 pKa = 7.84MTDD382 pKa = 3.49KK383 pKa = 10.69YY384 pKa = 11.15NQVHH388 pKa = 7.18DD389 pKa = 4.28EE390 pKa = 4.27LSAQVSDD397 pKa = 4.41AFIAQISPRR406 pKa = 11.84NSPTNLHH413 pKa = 6.35ILL415 pKa = 3.73
Molecular weight: 48.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
11191 |
38 |
1429 |
243.3 |
27.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.087 ± 0.639 | 0.608 ± 0.116 |
5.987 ± 0.371 | 7.819 ± 0.328 |
3.628 ± 0.313 | 6.738 ± 0.329 |
1.287 ± 0.095 | 5.987 ± 0.257 |
8.435 ± 0.277 | 8.283 ± 0.343 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.359 ± 0.162 | 5.719 ± 0.281 |
3.101 ± 0.218 | 4.218 ± 0.251 |
4.298 ± 0.282 | 6.05 ± 0.317 |
5.987 ± 0.308 | 6.148 ± 0.258 |
1.394 ± 0.169 | 3.869 ± 0.336 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
