
Roseibium hamelinense
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Stappiaceae; Roseibium
Average proteome isoelectric point is 6.16
Get precalculated fractions of proteins
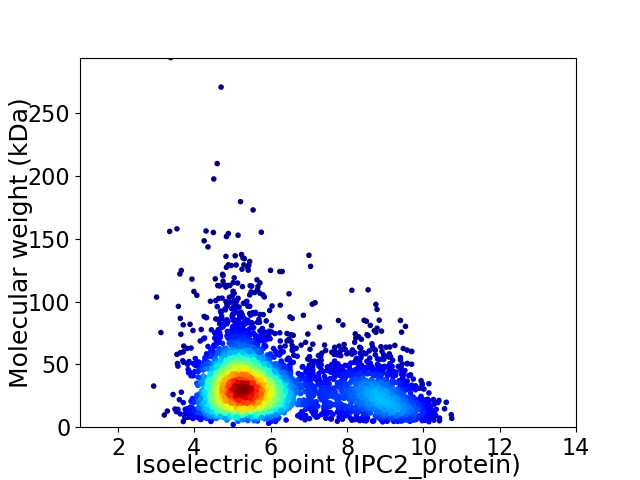
Virtual 2D-PAGE plot for 4355 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A562T0Y1|A0A562T0Y1_9RHOB Uncharacterized protein OS=Roseibium hamelinense OX=150831 GN=JM93_02471 PE=4 SV=1
MM1 pKa = 7.7PNIFEE6 pKa = 4.34TSDD9 pKa = 3.41AASGTSTSYY18 pKa = 11.32VLRR21 pKa = 11.84EE22 pKa = 3.95TGKK25 pKa = 10.09IAQGTQNAVFTTVQAHH41 pKa = 6.92LAPGDD46 pKa = 3.83DD47 pKa = 4.17SDD49 pKa = 4.67WFAIDD54 pKa = 4.31LKK56 pKa = 11.1AGQTYY61 pKa = 8.14TFAAVGTFDD70 pKa = 3.66GRR72 pKa = 11.84GRR74 pKa = 11.84LEE76 pKa = 4.34DD77 pKa = 4.73PYY79 pKa = 11.56LKK81 pKa = 10.91LLDD84 pKa = 3.83QSGSLIGEE92 pKa = 4.74DD93 pKa = 4.48DD94 pKa = 3.93DD95 pKa = 5.56AGPGAYY101 pKa = 9.76SAITFTAQTSGRR113 pKa = 11.84YY114 pKa = 8.3FLSVSAGEE122 pKa = 4.36AGLTGQYY129 pKa = 10.68GLSAVEE135 pKa = 4.1GTKK138 pKa = 10.41AVYY141 pKa = 10.4DD142 pKa = 3.67GDD144 pKa = 3.95MAAGALLRR152 pKa = 11.84PGVSWGTPGQPATVTYY168 pKa = 10.2GFRR171 pKa = 11.84DD172 pKa = 3.57TLNNLDD178 pKa = 3.91PEE180 pKa = 4.53YY181 pKa = 10.58AAEE184 pKa = 4.08VPEE187 pKa = 4.23TFEE190 pKa = 4.35QVTAAQMVAVEE201 pKa = 5.57AILATYY207 pKa = 10.37SEE209 pKa = 4.16IANIQFEE216 pKa = 4.53RR217 pKa = 11.84VNPDD221 pKa = 2.86GYY223 pKa = 9.63TDD225 pKa = 3.86DD226 pKa = 5.58ASMLFSNYY234 pKa = 9.14FSTTDD239 pKa = 3.21GAGAYY244 pKa = 9.84AYY246 pKa = 10.42FPGSDD251 pKa = 3.77ASDD254 pKa = 3.02RR255 pKa = 11.84WEE257 pKa = 4.23GDD259 pKa = 2.52VWLNTNSVSKK269 pKa = 11.14EE270 pKa = 3.71EE271 pKa = 4.59LPNGSYY277 pKa = 10.22SHH279 pKa = 6.84FAVMHH284 pKa = 6.39EE285 pKa = 4.2IGHH288 pKa = 6.23AVGLHH293 pKa = 6.24HH294 pKa = 7.29PGDD297 pKa = 4.36YY298 pKa = 10.35NASPDD303 pKa = 3.29IDD305 pKa = 3.24ITYY308 pKa = 10.3EE309 pKa = 3.56AFAQYY314 pKa = 10.66RR315 pKa = 11.84EE316 pKa = 4.26DD317 pKa = 3.73TQQYY321 pKa = 10.8SVMSYY326 pKa = 10.69FDD328 pKa = 3.7EE329 pKa = 4.8SNADD333 pKa = 3.19GSLGFGAFPDD343 pKa = 3.88TLMLHH348 pKa = 7.44DD349 pKa = 5.2ILALQQLYY357 pKa = 9.93GANTSTRR364 pKa = 11.84SGDD367 pKa = 3.08TTYY370 pKa = 11.36GFNSTEE376 pKa = 3.73GGVYY380 pKa = 10.38DD381 pKa = 5.25FSQAMPHH388 pKa = 7.27ALTLWDD394 pKa = 4.02AGGVDD399 pKa = 4.41TIDD402 pKa = 3.45LSGAGKK408 pKa = 10.09VDD410 pKa = 3.49QFVSLIDD417 pKa = 3.55GTYY420 pKa = 9.68SNVGGGLGNMAIAYY434 pKa = 8.57SAVIEE439 pKa = 4.07NAIGGRR445 pKa = 11.84GDD447 pKa = 4.7DD448 pKa = 3.58VVLGNAASNTLHH460 pKa = 6.75GKK462 pKa = 10.18AGDD465 pKa = 3.75DD466 pKa = 3.78TLVGGAGHH474 pKa = 7.45DD475 pKa = 3.67KK476 pKa = 11.2LFGGSGNDD484 pKa = 3.31RR485 pKa = 11.84LDD487 pKa = 3.88GGSSDD492 pKa = 5.02DD493 pKa = 4.02VLNGGAGDD501 pKa = 4.18DD502 pKa = 4.01SLIGGSGNDD511 pKa = 3.15RR512 pKa = 11.84LVGGSGNDD520 pKa = 3.46TYY522 pKa = 11.27TGGSGSDD529 pKa = 3.11MFCIYY534 pKa = 10.78NRR536 pKa = 11.84GDD538 pKa = 3.42AEE540 pKa = 4.69TITDD544 pKa = 4.12FQSGFDD550 pKa = 3.76TIAIRR555 pKa = 11.84SEE557 pKa = 3.78FAAFDD562 pKa = 3.8NLTILASGADD572 pKa = 3.79SVIKK576 pKa = 10.74FGDD579 pKa = 3.44NSISLLGVDD588 pKa = 5.19AGIISQNDD596 pKa = 3.45LAFVV600 pKa = 3.67
MM1 pKa = 7.7PNIFEE6 pKa = 4.34TSDD9 pKa = 3.41AASGTSTSYY18 pKa = 11.32VLRR21 pKa = 11.84EE22 pKa = 3.95TGKK25 pKa = 10.09IAQGTQNAVFTTVQAHH41 pKa = 6.92LAPGDD46 pKa = 3.83DD47 pKa = 4.17SDD49 pKa = 4.67WFAIDD54 pKa = 4.31LKK56 pKa = 11.1AGQTYY61 pKa = 8.14TFAAVGTFDD70 pKa = 3.66GRR72 pKa = 11.84GRR74 pKa = 11.84LEE76 pKa = 4.34DD77 pKa = 4.73PYY79 pKa = 11.56LKK81 pKa = 10.91LLDD84 pKa = 3.83QSGSLIGEE92 pKa = 4.74DD93 pKa = 4.48DD94 pKa = 3.93DD95 pKa = 5.56AGPGAYY101 pKa = 9.76SAITFTAQTSGRR113 pKa = 11.84YY114 pKa = 8.3FLSVSAGEE122 pKa = 4.36AGLTGQYY129 pKa = 10.68GLSAVEE135 pKa = 4.1GTKK138 pKa = 10.41AVYY141 pKa = 10.4DD142 pKa = 3.67GDD144 pKa = 3.95MAAGALLRR152 pKa = 11.84PGVSWGTPGQPATVTYY168 pKa = 10.2GFRR171 pKa = 11.84DD172 pKa = 3.57TLNNLDD178 pKa = 3.91PEE180 pKa = 4.53YY181 pKa = 10.58AAEE184 pKa = 4.08VPEE187 pKa = 4.23TFEE190 pKa = 4.35QVTAAQMVAVEE201 pKa = 5.57AILATYY207 pKa = 10.37SEE209 pKa = 4.16IANIQFEE216 pKa = 4.53RR217 pKa = 11.84VNPDD221 pKa = 2.86GYY223 pKa = 9.63TDD225 pKa = 3.86DD226 pKa = 5.58ASMLFSNYY234 pKa = 9.14FSTTDD239 pKa = 3.21GAGAYY244 pKa = 9.84AYY246 pKa = 10.42FPGSDD251 pKa = 3.77ASDD254 pKa = 3.02RR255 pKa = 11.84WEE257 pKa = 4.23GDD259 pKa = 2.52VWLNTNSVSKK269 pKa = 11.14EE270 pKa = 3.71EE271 pKa = 4.59LPNGSYY277 pKa = 10.22SHH279 pKa = 6.84FAVMHH284 pKa = 6.39EE285 pKa = 4.2IGHH288 pKa = 6.23AVGLHH293 pKa = 6.24HH294 pKa = 7.29PGDD297 pKa = 4.36YY298 pKa = 10.35NASPDD303 pKa = 3.29IDD305 pKa = 3.24ITYY308 pKa = 10.3EE309 pKa = 3.56AFAQYY314 pKa = 10.66RR315 pKa = 11.84EE316 pKa = 4.26DD317 pKa = 3.73TQQYY321 pKa = 10.8SVMSYY326 pKa = 10.69FDD328 pKa = 3.7EE329 pKa = 4.8SNADD333 pKa = 3.19GSLGFGAFPDD343 pKa = 3.88TLMLHH348 pKa = 7.44DD349 pKa = 5.2ILALQQLYY357 pKa = 9.93GANTSTRR364 pKa = 11.84SGDD367 pKa = 3.08TTYY370 pKa = 11.36GFNSTEE376 pKa = 3.73GGVYY380 pKa = 10.38DD381 pKa = 5.25FSQAMPHH388 pKa = 7.27ALTLWDD394 pKa = 4.02AGGVDD399 pKa = 4.41TIDD402 pKa = 3.45LSGAGKK408 pKa = 10.09VDD410 pKa = 3.49QFVSLIDD417 pKa = 3.55GTYY420 pKa = 9.68SNVGGGLGNMAIAYY434 pKa = 8.57SAVIEE439 pKa = 4.07NAIGGRR445 pKa = 11.84GDD447 pKa = 4.7DD448 pKa = 3.58VVLGNAASNTLHH460 pKa = 6.75GKK462 pKa = 10.18AGDD465 pKa = 3.75DD466 pKa = 3.78TLVGGAGHH474 pKa = 7.45DD475 pKa = 3.67KK476 pKa = 11.2LFGGSGNDD484 pKa = 3.31RR485 pKa = 11.84LDD487 pKa = 3.88GGSSDD492 pKa = 5.02DD493 pKa = 4.02VLNGGAGDD501 pKa = 4.18DD502 pKa = 4.01SLIGGSGNDD511 pKa = 3.15RR512 pKa = 11.84LVGGSGNDD520 pKa = 3.46TYY522 pKa = 11.27TGGSGSDD529 pKa = 3.11MFCIYY534 pKa = 10.78NRR536 pKa = 11.84GDD538 pKa = 3.42AEE540 pKa = 4.69TITDD544 pKa = 4.12FQSGFDD550 pKa = 3.76TIAIRR555 pKa = 11.84SEE557 pKa = 3.78FAAFDD562 pKa = 3.8NLTILASGADD572 pKa = 3.79SVIKK576 pKa = 10.74FGDD579 pKa = 3.44NSISLLGVDD588 pKa = 5.19AGIISQNDD596 pKa = 3.45LAFVV600 pKa = 3.67
Molecular weight: 62.67 kDa
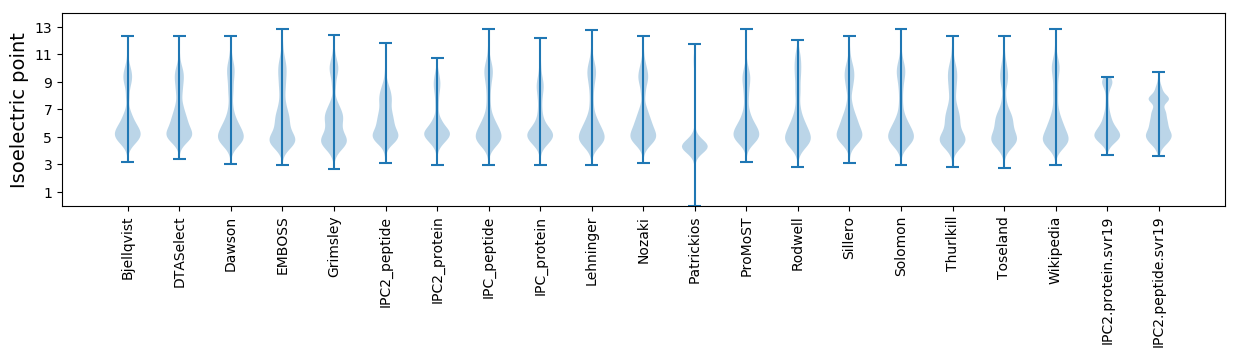
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A562SLY9|A0A562SLY9_9RHOB Probable membrane transporter protein OS=Roseibium hamelinense OX=150831 GN=JM93_03579 PE=3 SV=1
MM1 pKa = 7.36GHH3 pKa = 7.46LFWLSDD9 pKa = 3.78EE10 pKa = 3.92QWAAIEE16 pKa = 4.11PLLPRR21 pKa = 11.84NKK23 pKa = 10.16GGARR27 pKa = 11.84RR28 pKa = 11.84VDD30 pKa = 3.5DD31 pKa = 3.8RR32 pKa = 11.84RR33 pKa = 11.84VISGIIHH40 pKa = 6.07GLKK43 pKa = 10.18VGCRR47 pKa = 11.84WCDD50 pKa = 3.36CPEE53 pKa = 4.02EE54 pKa = 4.37YY55 pKa = 10.87GPSTTIYY62 pKa = 10.63NRR64 pKa = 11.84FNRR67 pKa = 11.84WSRR70 pKa = 11.84KK71 pKa = 9.35RR72 pKa = 11.84FWIDD76 pKa = 3.16LVEE79 pKa = 4.26GLAICGAVTKK89 pKa = 9.64STSIDD94 pKa = 3.22STYY97 pKa = 11.31VKK99 pKa = 10.34AHH101 pKa = 6.76RR102 pKa = 11.84SAHH105 pKa = 5.41GGKK108 pKa = 10.02GGRR111 pKa = 11.84KK112 pKa = 7.96IRR114 pKa = 11.84LSGRR118 pKa = 11.84HH119 pKa = 4.87GAGKK123 pKa = 10.06RR124 pKa = 11.84RR125 pKa = 11.84RR126 pKa = 11.84STPLPTT132 pKa = 3.9
MM1 pKa = 7.36GHH3 pKa = 7.46LFWLSDD9 pKa = 3.78EE10 pKa = 3.92QWAAIEE16 pKa = 4.11PLLPRR21 pKa = 11.84NKK23 pKa = 10.16GGARR27 pKa = 11.84RR28 pKa = 11.84VDD30 pKa = 3.5DD31 pKa = 3.8RR32 pKa = 11.84RR33 pKa = 11.84VISGIIHH40 pKa = 6.07GLKK43 pKa = 10.18VGCRR47 pKa = 11.84WCDD50 pKa = 3.36CPEE53 pKa = 4.02EE54 pKa = 4.37YY55 pKa = 10.87GPSTTIYY62 pKa = 10.63NRR64 pKa = 11.84FNRR67 pKa = 11.84WSRR70 pKa = 11.84KK71 pKa = 9.35RR72 pKa = 11.84FWIDD76 pKa = 3.16LVEE79 pKa = 4.26GLAICGAVTKK89 pKa = 9.64STSIDD94 pKa = 3.22STYY97 pKa = 11.31VKK99 pKa = 10.34AHH101 pKa = 6.76RR102 pKa = 11.84SAHH105 pKa = 5.41GGKK108 pKa = 10.02GGRR111 pKa = 11.84KK112 pKa = 7.96IRR114 pKa = 11.84LSGRR118 pKa = 11.84HH119 pKa = 4.87GAGKK123 pKa = 10.06RR124 pKa = 11.84RR125 pKa = 11.84RR126 pKa = 11.84STPLPTT132 pKa = 3.9
Molecular weight: 14.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1400321 |
21 |
2801 |
321.5 |
35.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.299 ± 0.046 | 0.889 ± 0.011 |
6.017 ± 0.041 | 6.003 ± 0.037 |
4.015 ± 0.024 | 8.272 ± 0.035 |
2.007 ± 0.02 | 5.387 ± 0.026 |
3.897 ± 0.028 | 10.007 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.479 ± 0.018 | 3.055 ± 0.024 |
4.874 ± 0.03 | 3.395 ± 0.021 |
6.095 ± 0.037 | 5.958 ± 0.031 |
5.531 ± 0.03 | 7.26 ± 0.03 |
1.239 ± 0.014 | 2.322 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
