
Rachicladosporium antarcticum
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; dothideomyceta; Dothideomycetes; Dothideomycetidae; Cladosporiales; Cladosporiaceae; Rachicladosporium
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins
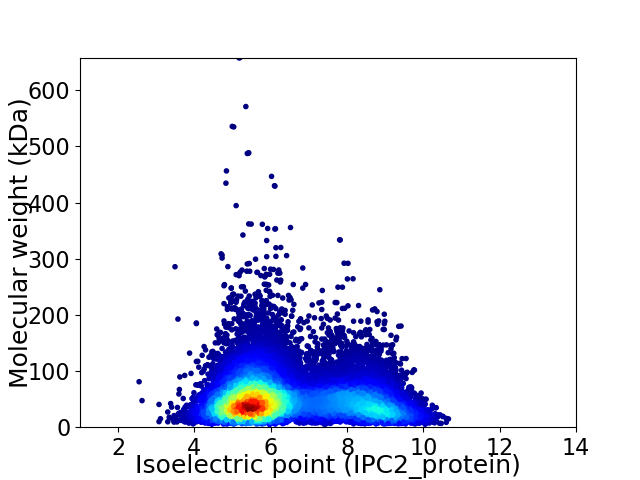
Virtual 2D-PAGE plot for 18598 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1V8SNQ6|A0A1V8SNQ6_9PEZI PX domain-containing protein OS=Rachicladosporium antarcticum OX=1507870 GN=B0A48_13872 PE=4 SV=1
MM1 pKa = 7.81ASPSTILTALLAATTTTSPLPAQTFGPQTVNLVSRR36 pKa = 11.84LCADD40 pKa = 3.22QSTSCSDD47 pKa = 3.24TTYY50 pKa = 10.86LAWLNTPFSLNITTSQLSLSSAQNVDD76 pKa = 2.8IDD78 pKa = 4.3LVNCEE83 pKa = 4.09VFFTGGDD90 pKa = 3.36AGPLLTADD98 pKa = 3.89TPVTLSSGLVDD109 pKa = 3.99IGNVTCSVSDD119 pKa = 3.38EE120 pKa = 4.79GKK122 pKa = 10.56GQDD125 pKa = 2.92QAGQIRR131 pKa = 11.84PGDD134 pKa = 3.62NATTTWVHH142 pKa = 6.14GGDD145 pKa = 4.89AIPVTTTTATGAVPTITGWGGAVVDD170 pKa = 4.92GGNEE174 pKa = 3.86TTATGSTPPSTTTYY188 pKa = 9.81TGGASGLEE196 pKa = 4.28VARR199 pKa = 11.84LLAAIAAVVGGWVSLLL215 pKa = 3.61
MM1 pKa = 7.81ASPSTILTALLAATTTTSPLPAQTFGPQTVNLVSRR36 pKa = 11.84LCADD40 pKa = 3.22QSTSCSDD47 pKa = 3.24TTYY50 pKa = 10.86LAWLNTPFSLNITTSQLSLSSAQNVDD76 pKa = 2.8IDD78 pKa = 4.3LVNCEE83 pKa = 4.09VFFTGGDD90 pKa = 3.36AGPLLTADD98 pKa = 3.89TPVTLSSGLVDD109 pKa = 3.99IGNVTCSVSDD119 pKa = 3.38EE120 pKa = 4.79GKK122 pKa = 10.56GQDD125 pKa = 2.92QAGQIRR131 pKa = 11.84PGDD134 pKa = 3.62NATTTWVHH142 pKa = 6.14GGDD145 pKa = 4.89AIPVTTTTATGAVPTITGWGGAVVDD170 pKa = 4.92GGNEE174 pKa = 3.86TTATGSTPPSTTTYY188 pKa = 9.81TGGASGLEE196 pKa = 4.28VARR199 pKa = 11.84LLAAIAAVVGGWVSLLL215 pKa = 3.61
Molecular weight: 21.51 kDa
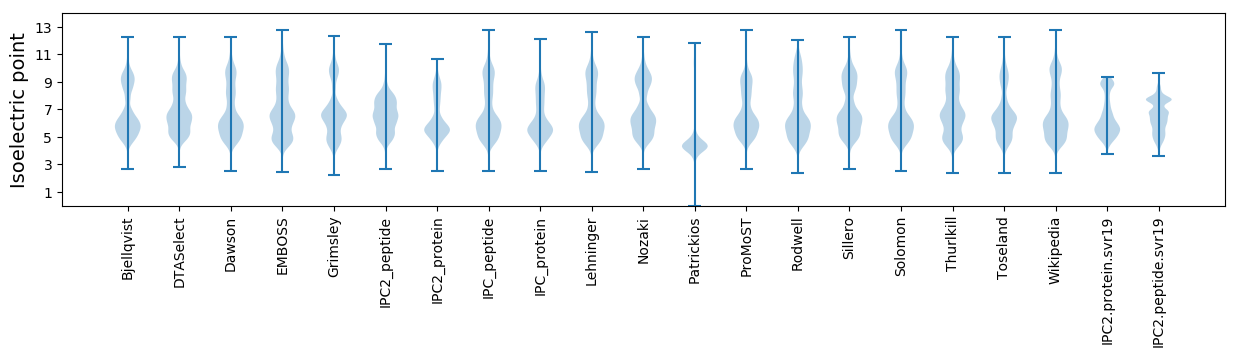
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1V8T4W8|A0A1V8T4W8_9PEZI 60S acidic ribosomal protein P2 OS=Rachicladosporium antarcticum OX=1507870 GN=B0A48_08948 PE=3 SV=1
MM1 pKa = 7.29NGDD4 pKa = 3.64SGKK7 pKa = 10.74LPLIDD12 pKa = 4.36IGDD15 pKa = 3.82EE16 pKa = 3.88ARR18 pKa = 11.84VCRR21 pKa = 11.84RR22 pKa = 11.84ANAARR27 pKa = 11.84PTDD30 pKa = 3.6NPARR34 pKa = 11.84TLLKK38 pKa = 9.92EE39 pKa = 3.96AYY41 pKa = 8.96KK42 pKa = 11.05AEE44 pKa = 4.01LAEE47 pKa = 4.52LGIDD51 pKa = 3.58PGQDD55 pKa = 2.3RR56 pKa = 11.84VIFDD60 pKa = 4.34CLWNIAHH67 pKa = 6.25VVTEE71 pKa = 4.34KK72 pKa = 10.08WRR74 pKa = 11.84STGAVDD80 pKa = 4.35FLEE83 pKa = 5.05ALAQEE88 pKa = 4.49FDD90 pKa = 3.17KK91 pKa = 11.4HH92 pKa = 6.41GITIVEE98 pKa = 4.29HH99 pKa = 7.45DD100 pKa = 4.6DD101 pKa = 4.01DD102 pKa = 4.2GTEE105 pKa = 4.0ITHH108 pKa = 6.5EE109 pKa = 4.31APVQQVVSSRR119 pKa = 11.84KK120 pKa = 9.25QIVDD124 pKa = 3.05KK125 pKa = 11.05RR126 pKa = 11.84RR127 pKa = 11.84VSFDD131 pKa = 3.34DD132 pKa = 3.89ARR134 pKa = 11.84MHH136 pKa = 5.51EE137 pKa = 4.01TWIRR141 pKa = 11.84EE142 pKa = 3.95RR143 pKa = 11.84TEE145 pKa = 3.81TLTQQEE151 pKa = 4.37DD152 pKa = 3.82APARR156 pKa = 11.84LLSQPPVRR164 pKa = 11.84ASLQRR169 pKa = 11.84RR170 pKa = 11.84ARR172 pKa = 11.84SVSSEE177 pKa = 3.88HH178 pKa = 7.14GYY180 pKa = 8.4ATYY183 pKa = 10.55QPGRR187 pKa = 11.84GASLYY192 pKa = 10.45GRR194 pKa = 11.84PQSSTDD200 pKa = 3.39QASDD204 pKa = 3.46PDD206 pKa = 4.23GPGDD210 pKa = 3.49SSMLRR215 pKa = 11.84RR216 pKa = 11.84LEE218 pKa = 3.86VSFEE222 pKa = 4.34KK223 pKa = 10.53IATLQEE229 pKa = 3.83LRR231 pKa = 11.84EE232 pKa = 4.4TCEE235 pKa = 3.6HH236 pKa = 5.44TRR238 pKa = 11.84EE239 pKa = 3.88LRR241 pKa = 11.84LVNTYY246 pKa = 9.14FNRR249 pKa = 11.84WFGNTHH255 pKa = 6.44LHH257 pKa = 4.7QQHH260 pKa = 5.89VARR263 pKa = 11.84QHH265 pKa = 6.22SEE267 pKa = 3.97AEE269 pKa = 4.03EE270 pKa = 3.69QHH272 pKa = 6.98AKK274 pKa = 10.24VLCQAVFDD282 pKa = 3.94SWRR285 pKa = 11.84EE286 pKa = 3.82AFHH289 pKa = 6.69EE290 pKa = 4.07KK291 pKa = 10.33QILRR295 pKa = 11.84RR296 pKa = 11.84EE297 pKa = 4.0ALEE300 pKa = 4.17AEE302 pKa = 4.45EE303 pKa = 5.52AEE305 pKa = 4.78QEE307 pKa = 4.25RR308 pKa = 11.84QSQQQQEE315 pKa = 4.41DD316 pKa = 3.41EE317 pKa = 4.17RR318 pKa = 11.84MQRR321 pKa = 11.84LGQRR325 pKa = 11.84AEE327 pKa = 3.82KK328 pKa = 10.63CRR330 pKa = 11.84GKK332 pKa = 9.53MLLRR336 pKa = 11.84KK337 pKa = 9.45GLQHH341 pKa = 6.06WFARR345 pKa = 11.84HH346 pKa = 4.04EE347 pKa = 4.05VRR349 pKa = 11.84RR350 pKa = 11.84AEE352 pKa = 4.13LQRR355 pKa = 11.84ASKK358 pKa = 10.31FLLAMRR364 pKa = 11.84YY365 pKa = 7.15FRR367 pKa = 11.84RR368 pKa = 11.84WKK370 pKa = 10.52HH371 pKa = 5.06IALEE375 pKa = 4.24NASKK379 pKa = 10.66ARR381 pKa = 11.84SILTRR386 pKa = 11.84KK387 pKa = 9.47YY388 pKa = 8.63IRR390 pKa = 11.84VWRR393 pKa = 11.84EE394 pKa = 3.13RR395 pKa = 11.84TARR398 pKa = 11.84SHH400 pKa = 6.48LAEE403 pKa = 4.27EE404 pKa = 4.26QAAAHH409 pKa = 6.43HH410 pKa = 6.66EE411 pKa = 4.33EE412 pKa = 4.21TLQRR416 pKa = 11.84QCVKK420 pKa = 9.9RR421 pKa = 11.84WAKK424 pKa = 10.37VLFNIKK430 pKa = 10.04LEE432 pKa = 3.78KK433 pKa = 10.3RR434 pKa = 11.84RR435 pKa = 11.84IDD437 pKa = 4.08RR438 pKa = 11.84IKK440 pKa = 10.58HH441 pKa = 4.42AACRR445 pKa = 11.84KK446 pKa = 9.04WLDD449 pKa = 3.02AVQRR453 pKa = 11.84VRR455 pKa = 11.84ALEE458 pKa = 4.03AQAAAFRR465 pKa = 11.84SQRR468 pKa = 11.84LVRR471 pKa = 11.84PVLLALRR478 pKa = 11.84AGVLRR483 pKa = 11.84VQDD486 pKa = 3.89LASRR490 pKa = 11.84AQTCYY495 pKa = 10.31AQARR499 pKa = 11.84LTIAFSVMRR508 pKa = 11.84IQGRR512 pKa = 11.84LKK514 pKa = 10.03PLEE517 pKa = 3.92NVMTLRR523 pKa = 11.84LNLTLEE529 pKa = 4.38RR530 pKa = 11.84KK531 pKa = 9.65AFATWHH537 pKa = 6.03LQVQLAQQAAHH548 pKa = 6.68FDD550 pKa = 3.23QRR552 pKa = 11.84RR553 pKa = 11.84IKK555 pKa = 9.92RR556 pKa = 11.84AAWHH560 pKa = 6.0GWVDD564 pKa = 3.43ALKK567 pKa = 10.39CRR569 pKa = 11.84VVAQHH574 pKa = 5.58INARR578 pKa = 11.84VVIEE582 pKa = 4.18SLYY585 pKa = 10.72RR586 pKa = 11.84WMLQEE591 pKa = 3.98RR592 pKa = 11.84LQLFRR597 pKa = 11.84RR598 pKa = 11.84TADD601 pKa = 3.02ARR603 pKa = 11.84LLRR606 pKa = 11.84NSFAALKK613 pKa = 10.71GMSTDD618 pKa = 3.74LQAQLDD624 pKa = 3.7MRR626 pKa = 11.84AYY628 pKa = 10.59AFAQGQQRR636 pKa = 11.84RR637 pKa = 11.84RR638 pKa = 11.84LAGAMFRR645 pKa = 11.84LHH647 pKa = 5.37VLSRR651 pKa = 11.84RR652 pKa = 11.84RR653 pKa = 11.84EE654 pKa = 3.97DD655 pKa = 3.26AEE657 pKa = 3.88RR658 pKa = 11.84AAVEE662 pKa = 4.24FANSRR667 pKa = 11.84ALPAVLEE674 pKa = 4.36TWHH677 pKa = 6.79EE678 pKa = 4.32KK679 pKa = 6.36TTHH682 pKa = 4.0VTKK685 pKa = 10.82LNRR688 pKa = 11.84WATDD692 pKa = 2.64ARR694 pKa = 11.84FYY696 pKa = 10.45CLCSAALKK704 pKa = 9.68RR705 pKa = 11.84WRR707 pKa = 11.84DD708 pKa = 3.26ATVEE712 pKa = 4.08HH713 pKa = 5.09QHH715 pKa = 5.02QRR717 pKa = 11.84RR718 pKa = 11.84RR719 pKa = 11.84DD720 pKa = 3.11AYY722 pKa = 8.04AQVRR726 pKa = 11.84SRR728 pKa = 11.84YY729 pKa = 7.5KK730 pKa = 10.12HH731 pKa = 6.23RR732 pKa = 11.84LVGQCFASLQVKK744 pKa = 8.82YY745 pKa = 11.07AEE747 pKa = 4.49VYY749 pKa = 10.82SMSAAIQDD757 pKa = 3.81RR758 pKa = 11.84AQTTQTTTVAQLFATWRR775 pKa = 11.84SRR777 pKa = 11.84ARR779 pKa = 11.84TLQAQSLTASQLDD792 pKa = 3.75TSRR795 pKa = 11.84LLSSALSALTSQHH808 pKa = 6.09AQTTSLDD815 pKa = 3.69TEE817 pKa = 4.43AEE819 pKa = 4.21TFARR823 pKa = 11.84TTSLSLALSALKK835 pKa = 9.26TLQWATFTASQQSHH849 pKa = 5.21TALALRR855 pKa = 11.84SRR857 pKa = 11.84NRR859 pKa = 11.84EE860 pKa = 3.29QHH862 pKa = 5.03IRR864 pKa = 11.84HH865 pKa = 5.98MLRR868 pKa = 11.84HH869 pKa = 5.53WVSQTAARR877 pKa = 11.84RR878 pKa = 11.84AAATAEE884 pKa = 3.95PDD886 pKa = 3.49AEE888 pKa = 4.51LEE890 pKa = 4.42AEE892 pKa = 4.35PEE894 pKa = 4.4SPSLRR899 pKa = 11.84PASRR903 pKa = 11.84AAARR907 pKa = 11.84SLSRR911 pKa = 11.84DD912 pKa = 3.69RR913 pKa = 11.84IRR915 pKa = 11.84DD916 pKa = 3.37RR917 pKa = 11.84YY918 pKa = 8.4TSPTRR923 pKa = 11.84LPQGYY928 pKa = 9.96AGSTPGYY935 pKa = 9.68LRR937 pKa = 11.84TPSRR941 pKa = 11.84SRR943 pKa = 11.84RR944 pKa = 11.84AVGRR948 pKa = 11.84FRR950 pKa = 11.84PIPTPGPYY958 pKa = 9.31TPLAFTPAYY967 pKa = 9.8LVTGIPGVQVDD978 pKa = 4.9DD979 pKa = 4.55GAGLDD984 pKa = 4.01MGSSPPLPMALAGSGQVTPFAQRR1007 pKa = 11.84LRR1009 pKa = 11.84EE1010 pKa = 4.11VGVVEE1015 pKa = 5.68AIPTGAITGAGAEE1028 pKa = 4.0FRR1030 pKa = 11.84PVSVLRR1036 pKa = 11.84RR1037 pKa = 11.84GLRR1040 pKa = 11.84QSVTGGTGTGKK1051 pKa = 9.69SVRR1054 pKa = 11.84FAGAGRR1060 pKa = 11.84FSSVGGGSRR1069 pKa = 11.84GQEE1072 pKa = 3.91HH1073 pKa = 6.94LKK1075 pKa = 10.97SSS1077 pKa = 3.56
MM1 pKa = 7.29NGDD4 pKa = 3.64SGKK7 pKa = 10.74LPLIDD12 pKa = 4.36IGDD15 pKa = 3.82EE16 pKa = 3.88ARR18 pKa = 11.84VCRR21 pKa = 11.84RR22 pKa = 11.84ANAARR27 pKa = 11.84PTDD30 pKa = 3.6NPARR34 pKa = 11.84TLLKK38 pKa = 9.92EE39 pKa = 3.96AYY41 pKa = 8.96KK42 pKa = 11.05AEE44 pKa = 4.01LAEE47 pKa = 4.52LGIDD51 pKa = 3.58PGQDD55 pKa = 2.3RR56 pKa = 11.84VIFDD60 pKa = 4.34CLWNIAHH67 pKa = 6.25VVTEE71 pKa = 4.34KK72 pKa = 10.08WRR74 pKa = 11.84STGAVDD80 pKa = 4.35FLEE83 pKa = 5.05ALAQEE88 pKa = 4.49FDD90 pKa = 3.17KK91 pKa = 11.4HH92 pKa = 6.41GITIVEE98 pKa = 4.29HH99 pKa = 7.45DD100 pKa = 4.6DD101 pKa = 4.01DD102 pKa = 4.2GTEE105 pKa = 4.0ITHH108 pKa = 6.5EE109 pKa = 4.31APVQQVVSSRR119 pKa = 11.84KK120 pKa = 9.25QIVDD124 pKa = 3.05KK125 pKa = 11.05RR126 pKa = 11.84RR127 pKa = 11.84VSFDD131 pKa = 3.34DD132 pKa = 3.89ARR134 pKa = 11.84MHH136 pKa = 5.51EE137 pKa = 4.01TWIRR141 pKa = 11.84EE142 pKa = 3.95RR143 pKa = 11.84TEE145 pKa = 3.81TLTQQEE151 pKa = 4.37DD152 pKa = 3.82APARR156 pKa = 11.84LLSQPPVRR164 pKa = 11.84ASLQRR169 pKa = 11.84RR170 pKa = 11.84ARR172 pKa = 11.84SVSSEE177 pKa = 3.88HH178 pKa = 7.14GYY180 pKa = 8.4ATYY183 pKa = 10.55QPGRR187 pKa = 11.84GASLYY192 pKa = 10.45GRR194 pKa = 11.84PQSSTDD200 pKa = 3.39QASDD204 pKa = 3.46PDD206 pKa = 4.23GPGDD210 pKa = 3.49SSMLRR215 pKa = 11.84RR216 pKa = 11.84LEE218 pKa = 3.86VSFEE222 pKa = 4.34KK223 pKa = 10.53IATLQEE229 pKa = 3.83LRR231 pKa = 11.84EE232 pKa = 4.4TCEE235 pKa = 3.6HH236 pKa = 5.44TRR238 pKa = 11.84EE239 pKa = 3.88LRR241 pKa = 11.84LVNTYY246 pKa = 9.14FNRR249 pKa = 11.84WFGNTHH255 pKa = 6.44LHH257 pKa = 4.7QQHH260 pKa = 5.89VARR263 pKa = 11.84QHH265 pKa = 6.22SEE267 pKa = 3.97AEE269 pKa = 4.03EE270 pKa = 3.69QHH272 pKa = 6.98AKK274 pKa = 10.24VLCQAVFDD282 pKa = 3.94SWRR285 pKa = 11.84EE286 pKa = 3.82AFHH289 pKa = 6.69EE290 pKa = 4.07KK291 pKa = 10.33QILRR295 pKa = 11.84RR296 pKa = 11.84EE297 pKa = 4.0ALEE300 pKa = 4.17AEE302 pKa = 4.45EE303 pKa = 5.52AEE305 pKa = 4.78QEE307 pKa = 4.25RR308 pKa = 11.84QSQQQQEE315 pKa = 4.41DD316 pKa = 3.41EE317 pKa = 4.17RR318 pKa = 11.84MQRR321 pKa = 11.84LGQRR325 pKa = 11.84AEE327 pKa = 3.82KK328 pKa = 10.63CRR330 pKa = 11.84GKK332 pKa = 9.53MLLRR336 pKa = 11.84KK337 pKa = 9.45GLQHH341 pKa = 6.06WFARR345 pKa = 11.84HH346 pKa = 4.04EE347 pKa = 4.05VRR349 pKa = 11.84RR350 pKa = 11.84AEE352 pKa = 4.13LQRR355 pKa = 11.84ASKK358 pKa = 10.31FLLAMRR364 pKa = 11.84YY365 pKa = 7.15FRR367 pKa = 11.84RR368 pKa = 11.84WKK370 pKa = 10.52HH371 pKa = 5.06IALEE375 pKa = 4.24NASKK379 pKa = 10.66ARR381 pKa = 11.84SILTRR386 pKa = 11.84KK387 pKa = 9.47YY388 pKa = 8.63IRR390 pKa = 11.84VWRR393 pKa = 11.84EE394 pKa = 3.13RR395 pKa = 11.84TARR398 pKa = 11.84SHH400 pKa = 6.48LAEE403 pKa = 4.27EE404 pKa = 4.26QAAAHH409 pKa = 6.43HH410 pKa = 6.66EE411 pKa = 4.33EE412 pKa = 4.21TLQRR416 pKa = 11.84QCVKK420 pKa = 9.9RR421 pKa = 11.84WAKK424 pKa = 10.37VLFNIKK430 pKa = 10.04LEE432 pKa = 3.78KK433 pKa = 10.3RR434 pKa = 11.84RR435 pKa = 11.84IDD437 pKa = 4.08RR438 pKa = 11.84IKK440 pKa = 10.58HH441 pKa = 4.42AACRR445 pKa = 11.84KK446 pKa = 9.04WLDD449 pKa = 3.02AVQRR453 pKa = 11.84VRR455 pKa = 11.84ALEE458 pKa = 4.03AQAAAFRR465 pKa = 11.84SQRR468 pKa = 11.84LVRR471 pKa = 11.84PVLLALRR478 pKa = 11.84AGVLRR483 pKa = 11.84VQDD486 pKa = 3.89LASRR490 pKa = 11.84AQTCYY495 pKa = 10.31AQARR499 pKa = 11.84LTIAFSVMRR508 pKa = 11.84IQGRR512 pKa = 11.84LKK514 pKa = 10.03PLEE517 pKa = 3.92NVMTLRR523 pKa = 11.84LNLTLEE529 pKa = 4.38RR530 pKa = 11.84KK531 pKa = 9.65AFATWHH537 pKa = 6.03LQVQLAQQAAHH548 pKa = 6.68FDD550 pKa = 3.23QRR552 pKa = 11.84RR553 pKa = 11.84IKK555 pKa = 9.92RR556 pKa = 11.84AAWHH560 pKa = 6.0GWVDD564 pKa = 3.43ALKK567 pKa = 10.39CRR569 pKa = 11.84VVAQHH574 pKa = 5.58INARR578 pKa = 11.84VVIEE582 pKa = 4.18SLYY585 pKa = 10.72RR586 pKa = 11.84WMLQEE591 pKa = 3.98RR592 pKa = 11.84LQLFRR597 pKa = 11.84RR598 pKa = 11.84TADD601 pKa = 3.02ARR603 pKa = 11.84LLRR606 pKa = 11.84NSFAALKK613 pKa = 10.71GMSTDD618 pKa = 3.74LQAQLDD624 pKa = 3.7MRR626 pKa = 11.84AYY628 pKa = 10.59AFAQGQQRR636 pKa = 11.84RR637 pKa = 11.84RR638 pKa = 11.84LAGAMFRR645 pKa = 11.84LHH647 pKa = 5.37VLSRR651 pKa = 11.84RR652 pKa = 11.84RR653 pKa = 11.84EE654 pKa = 3.97DD655 pKa = 3.26AEE657 pKa = 3.88RR658 pKa = 11.84AAVEE662 pKa = 4.24FANSRR667 pKa = 11.84ALPAVLEE674 pKa = 4.36TWHH677 pKa = 6.79EE678 pKa = 4.32KK679 pKa = 6.36TTHH682 pKa = 4.0VTKK685 pKa = 10.82LNRR688 pKa = 11.84WATDD692 pKa = 2.64ARR694 pKa = 11.84FYY696 pKa = 10.45CLCSAALKK704 pKa = 9.68RR705 pKa = 11.84WRR707 pKa = 11.84DD708 pKa = 3.26ATVEE712 pKa = 4.08HH713 pKa = 5.09QHH715 pKa = 5.02QRR717 pKa = 11.84RR718 pKa = 11.84RR719 pKa = 11.84DD720 pKa = 3.11AYY722 pKa = 8.04AQVRR726 pKa = 11.84SRR728 pKa = 11.84YY729 pKa = 7.5KK730 pKa = 10.12HH731 pKa = 6.23RR732 pKa = 11.84LVGQCFASLQVKK744 pKa = 8.82YY745 pKa = 11.07AEE747 pKa = 4.49VYY749 pKa = 10.82SMSAAIQDD757 pKa = 3.81RR758 pKa = 11.84AQTTQTTTVAQLFATWRR775 pKa = 11.84SRR777 pKa = 11.84ARR779 pKa = 11.84TLQAQSLTASQLDD792 pKa = 3.75TSRR795 pKa = 11.84LLSSALSALTSQHH808 pKa = 6.09AQTTSLDD815 pKa = 3.69TEE817 pKa = 4.43AEE819 pKa = 4.21TFARR823 pKa = 11.84TTSLSLALSALKK835 pKa = 9.26TLQWATFTASQQSHH849 pKa = 5.21TALALRR855 pKa = 11.84SRR857 pKa = 11.84NRR859 pKa = 11.84EE860 pKa = 3.29QHH862 pKa = 5.03IRR864 pKa = 11.84HH865 pKa = 5.98MLRR868 pKa = 11.84HH869 pKa = 5.53WVSQTAARR877 pKa = 11.84RR878 pKa = 11.84AAATAEE884 pKa = 3.95PDD886 pKa = 3.49AEE888 pKa = 4.51LEE890 pKa = 4.42AEE892 pKa = 4.35PEE894 pKa = 4.4SPSLRR899 pKa = 11.84PASRR903 pKa = 11.84AAARR907 pKa = 11.84SLSRR911 pKa = 11.84DD912 pKa = 3.69RR913 pKa = 11.84IRR915 pKa = 11.84DD916 pKa = 3.37RR917 pKa = 11.84YY918 pKa = 8.4TSPTRR923 pKa = 11.84LPQGYY928 pKa = 9.96AGSTPGYY935 pKa = 9.68LRR937 pKa = 11.84TPSRR941 pKa = 11.84SRR943 pKa = 11.84RR944 pKa = 11.84AVGRR948 pKa = 11.84FRR950 pKa = 11.84PIPTPGPYY958 pKa = 9.31TPLAFTPAYY967 pKa = 9.8LVTGIPGVQVDD978 pKa = 4.9DD979 pKa = 4.55GAGLDD984 pKa = 4.01MGSSPPLPMALAGSGQVTPFAQRR1007 pKa = 11.84LRR1009 pKa = 11.84EE1010 pKa = 4.11VGVVEE1015 pKa = 5.68AIPTGAITGAGAEE1028 pKa = 4.0FRR1030 pKa = 11.84PVSVLRR1036 pKa = 11.84RR1037 pKa = 11.84GLRR1040 pKa = 11.84QSVTGGTGTGKK1051 pKa = 9.69SVRR1054 pKa = 11.84FAGAGRR1060 pKa = 11.84FSSVGGGSRR1069 pKa = 11.84GQEE1072 pKa = 3.91HH1073 pKa = 6.94LKK1075 pKa = 10.97SSS1077 pKa = 3.56
Molecular weight: 122.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
9390076 |
50 |
6082 |
504.9 |
55.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.694 ± 0.016 | 1.083 ± 0.006 |
5.769 ± 0.015 | 6.108 ± 0.017 |
3.367 ± 0.012 | 7.231 ± 0.018 |
2.384 ± 0.008 | 4.44 ± 0.014 |
4.701 ± 0.015 | 8.719 ± 0.02 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.202 ± 0.008 | 3.33 ± 0.01 |
6.047 ± 0.019 | 4.04 ± 0.013 |
6.198 ± 0.018 | 8.127 ± 0.018 |
6.293 ± 0.014 | 6.209 ± 0.013 |
1.392 ± 0.006 | 2.668 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
