
Turnip curly top virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Turncurtovirus
Average proteome isoelectric point is 7.55
Get precalculated fractions of proteins
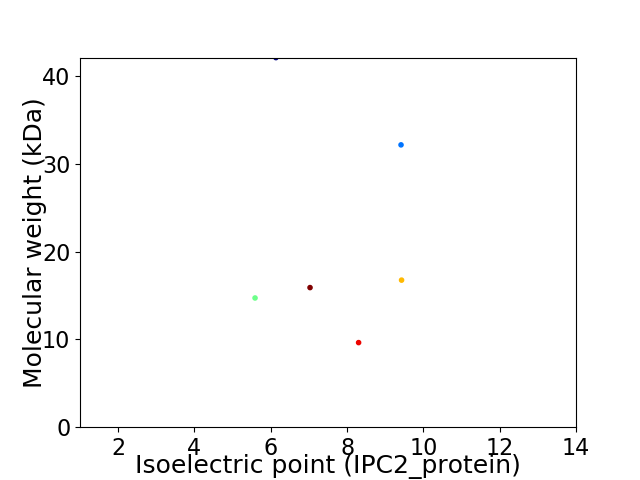
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|L0CSD0|L0CSD0_9GEMI Movement protein OS=Turnip curly top virus OX=859650 PE=4 SV=1
MM1 pKa = 7.42IWRR4 pKa = 11.84VNKK7 pKa = 10.0DD8 pKa = 3.35EE9 pKa = 4.01KK10 pKa = 10.39RR11 pKa = 11.84VRR13 pKa = 11.84AFRR16 pKa = 11.84YY17 pKa = 8.4KK18 pKa = 10.17KK19 pKa = 9.97RR20 pKa = 11.84RR21 pKa = 11.84FYY23 pKa = 11.51SLAEE27 pKa = 3.98YY28 pKa = 10.82LLAPLATHH36 pKa = 6.93LCKK39 pKa = 10.56ARR41 pKa = 11.84IFPDD45 pKa = 3.47DD46 pKa = 4.04KK47 pKa = 11.3YY48 pKa = 11.38LVPAYY53 pKa = 9.31TDD55 pKa = 3.2EE56 pKa = 5.01AYY58 pKa = 11.07SFLVFLKK65 pKa = 10.63EE66 pKa = 3.78EE67 pKa = 4.17TRR69 pKa = 11.84TPFQILKK76 pKa = 10.45DD77 pKa = 3.52EE78 pKa = 4.46WNVEE82 pKa = 4.15FKK84 pKa = 10.63EE85 pKa = 4.18IEE87 pKa = 4.16VEE89 pKa = 4.2LSRR92 pKa = 11.84LRR94 pKa = 11.84FDD96 pKa = 4.07PKK98 pKa = 11.14DD99 pKa = 3.77PGEE102 pKa = 4.15EE103 pKa = 4.15ACSEE107 pKa = 4.11EE108 pKa = 5.4SPVQQSSPVQSSSCEE123 pKa = 3.92TT124 pKa = 3.49
MM1 pKa = 7.42IWRR4 pKa = 11.84VNKK7 pKa = 10.0DD8 pKa = 3.35EE9 pKa = 4.01KK10 pKa = 10.39RR11 pKa = 11.84VRR13 pKa = 11.84AFRR16 pKa = 11.84YY17 pKa = 8.4KK18 pKa = 10.17KK19 pKa = 9.97RR20 pKa = 11.84RR21 pKa = 11.84FYY23 pKa = 11.51SLAEE27 pKa = 3.98YY28 pKa = 10.82LLAPLATHH36 pKa = 6.93LCKK39 pKa = 10.56ARR41 pKa = 11.84IFPDD45 pKa = 3.47DD46 pKa = 4.04KK47 pKa = 11.3YY48 pKa = 11.38LVPAYY53 pKa = 9.31TDD55 pKa = 3.2EE56 pKa = 5.01AYY58 pKa = 11.07SFLVFLKK65 pKa = 10.63EE66 pKa = 3.78EE67 pKa = 4.17TRR69 pKa = 11.84TPFQILKK76 pKa = 10.45DD77 pKa = 3.52EE78 pKa = 4.46WNVEE82 pKa = 4.15FKK84 pKa = 10.63EE85 pKa = 4.18IEE87 pKa = 4.16VEE89 pKa = 4.2LSRR92 pKa = 11.84LRR94 pKa = 11.84FDD96 pKa = 4.07PKK98 pKa = 11.14DD99 pKa = 3.77PGEE102 pKa = 4.15EE103 pKa = 4.15ACSEE107 pKa = 4.11EE108 pKa = 5.4SPVQQSSPVQSSSCEE123 pKa = 3.92TT124 pKa = 3.49
Molecular weight: 14.72 kDa
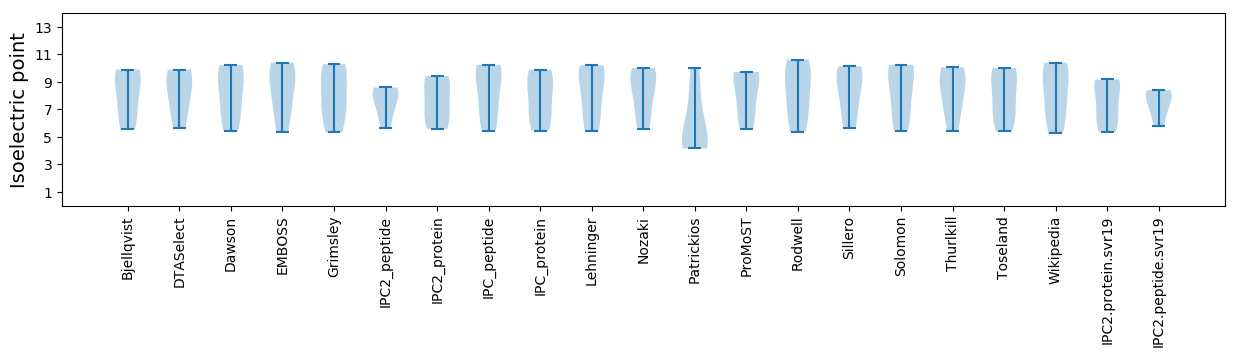
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|L0CRA1|L0CRA1_9GEMI Replication-associated protein OS=Turnip curly top virus OX=859650 PE=3 SV=1
MM1 pKa = 7.45SGTWSLKK8 pKa = 8.4RR9 pKa = 11.84SRR11 pKa = 11.84WNSPGYY17 pKa = 10.51DD18 pKa = 3.76LTPKK22 pKa = 10.03TPAKK26 pKa = 9.96RR27 pKa = 11.84PAVRR31 pKa = 11.84RR32 pKa = 11.84ALFNNQAQYY41 pKa = 11.39SRR43 pKa = 11.84VAARR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84WSKK52 pKa = 10.82VSVRR56 pKa = 11.84GNKK59 pKa = 9.29PYY61 pKa = 10.52RR62 pKa = 11.84SRR64 pKa = 11.84RR65 pKa = 11.84LKK67 pKa = 10.76ASDD70 pKa = 3.25YY71 pKa = 10.87QIYY74 pKa = 9.81KK75 pKa = 10.77DD76 pKa = 5.49KK77 pKa = 11.17IGGDD81 pKa = 3.69NGWTVTFSGDD91 pKa = 3.06CTMLNNYY98 pKa = 8.7VRR100 pKa = 11.84GIGRR104 pKa = 11.84DD105 pKa = 3.36QRR107 pKa = 11.84DD108 pKa = 3.3SVMTKK113 pKa = 7.87TAHH116 pKa = 4.22MHH118 pKa = 5.51FNGVLMANDD127 pKa = 4.52AFWEE131 pKa = 4.53APNYY135 pKa = 6.13MTMYY139 pKa = 10.36SWIILDD145 pKa = 4.08NDD147 pKa = 3.98PGGTFPKK154 pKa = 10.28PSDD157 pKa = 3.54IFDD160 pKa = 3.58MEE162 pKa = 4.48YY163 pKa = 10.97KK164 pKa = 9.98EE165 pKa = 4.46FPSMYY170 pKa = 9.81EE171 pKa = 3.57VAEE174 pKa = 4.3SVKK177 pKa = 10.65SRR179 pKa = 11.84FIVKK183 pKa = 10.19RR184 pKa = 11.84KK185 pKa = 7.01TEE187 pKa = 3.84HH188 pKa = 5.95YY189 pKa = 10.34LRR191 pKa = 11.84STGVAFGEE199 pKa = 4.24KK200 pKa = 9.68QNYY203 pKa = 7.41KK204 pKa = 10.24APNVGPIKK212 pKa = 10.58KK213 pKa = 9.24PIRR216 pKa = 11.84MKK218 pKa = 10.48FRR220 pKa = 11.84NIWQPSEE227 pKa = 3.97WKK229 pKa = 9.01DD230 pKa = 3.43TAGGKK235 pKa = 10.06YY236 pKa = 10.11EE237 pKa = 4.17DD238 pKa = 4.21LKK240 pKa = 11.25KK241 pKa = 10.83GALLYY246 pKa = 11.03VCICDD251 pKa = 3.71NKK253 pKa = 9.7ATQFSFNLKK262 pKa = 9.68GQWTMYY268 pKa = 10.19FINRR272 pKa = 11.84DD273 pKa = 3.02LMYY276 pKa = 11.06
MM1 pKa = 7.45SGTWSLKK8 pKa = 8.4RR9 pKa = 11.84SRR11 pKa = 11.84WNSPGYY17 pKa = 10.51DD18 pKa = 3.76LTPKK22 pKa = 10.03TPAKK26 pKa = 9.96RR27 pKa = 11.84PAVRR31 pKa = 11.84RR32 pKa = 11.84ALFNNQAQYY41 pKa = 11.39SRR43 pKa = 11.84VAARR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84WSKK52 pKa = 10.82VSVRR56 pKa = 11.84GNKK59 pKa = 9.29PYY61 pKa = 10.52RR62 pKa = 11.84SRR64 pKa = 11.84RR65 pKa = 11.84LKK67 pKa = 10.76ASDD70 pKa = 3.25YY71 pKa = 10.87QIYY74 pKa = 9.81KK75 pKa = 10.77DD76 pKa = 5.49KK77 pKa = 11.17IGGDD81 pKa = 3.69NGWTVTFSGDD91 pKa = 3.06CTMLNNYY98 pKa = 8.7VRR100 pKa = 11.84GIGRR104 pKa = 11.84DD105 pKa = 3.36QRR107 pKa = 11.84DD108 pKa = 3.3SVMTKK113 pKa = 7.87TAHH116 pKa = 4.22MHH118 pKa = 5.51FNGVLMANDD127 pKa = 4.52AFWEE131 pKa = 4.53APNYY135 pKa = 6.13MTMYY139 pKa = 10.36SWIILDD145 pKa = 4.08NDD147 pKa = 3.98PGGTFPKK154 pKa = 10.28PSDD157 pKa = 3.54IFDD160 pKa = 3.58MEE162 pKa = 4.48YY163 pKa = 10.97KK164 pKa = 9.98EE165 pKa = 4.46FPSMYY170 pKa = 9.81EE171 pKa = 3.57VAEE174 pKa = 4.3SVKK177 pKa = 10.65SRR179 pKa = 11.84FIVKK183 pKa = 10.19RR184 pKa = 11.84KK185 pKa = 7.01TEE187 pKa = 3.84HH188 pKa = 5.95YY189 pKa = 10.34LRR191 pKa = 11.84STGVAFGEE199 pKa = 4.24KK200 pKa = 9.68QNYY203 pKa = 7.41KK204 pKa = 10.24APNVGPIKK212 pKa = 10.58KK213 pKa = 9.24PIRR216 pKa = 11.84MKK218 pKa = 10.48FRR220 pKa = 11.84NIWQPSEE227 pKa = 3.97WKK229 pKa = 9.01DD230 pKa = 3.43TAGGKK235 pKa = 10.06YY236 pKa = 10.11EE237 pKa = 4.17DD238 pKa = 4.21LKK240 pKa = 11.25KK241 pKa = 10.83GALLYY246 pKa = 11.03VCICDD251 pKa = 3.71NKK253 pKa = 9.7ATQFSFNLKK262 pKa = 9.68GQWTMYY268 pKa = 10.19FINRR272 pKa = 11.84DD273 pKa = 3.02LMYY276 pKa = 11.06
Molecular weight: 32.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1138 |
85 |
371 |
189.7 |
21.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.921 ± 0.675 | 1.406 ± 0.224 |
5.097 ± 0.697 | 5.8 ± 0.938 |
4.833 ± 0.685 | 5.448 ± 0.723 |
3.427 ± 0.856 | 7.03 ± 1.027 |
6.678 ± 0.843 | 7.557 ± 0.897 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.285 ± 0.855 | 5.097 ± 0.652 |
5.8 ± 0.618 | 4.13 ± 0.426 |
6.415 ± 0.892 | 7.909 ± 1.077 |
5.536 ± 0.52 | 4.745 ± 0.576 |
1.845 ± 0.398 | 4.042 ± 0.663 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
