
Lake Sarah-associated circular virus-1
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 8.3
Get precalculated fractions of proteins
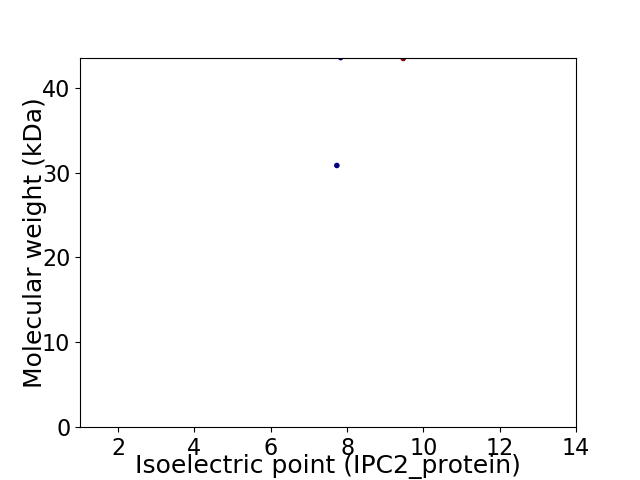
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
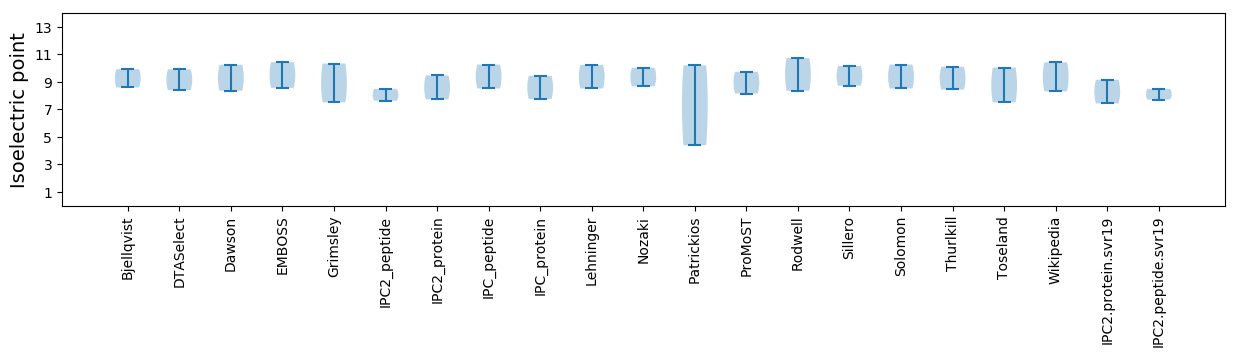
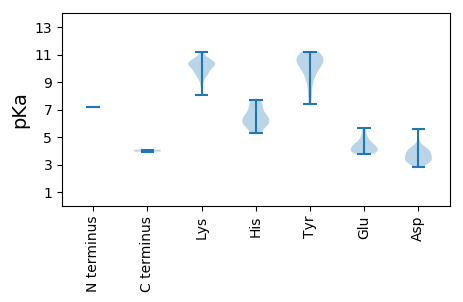
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140AQJ3|A0A140AQJ3_9VIRU Replication associated protein OS=Lake Sarah-associated circular virus-1 OX=1685735 PE=4 SV=1
MM1 pKa = 7.16SQGIYY6 pKa = 9.63WLLTINQHH14 pKa = 5.28EE15 pKa = 4.51FVPYY19 pKa = 10.13LPAGVSYY26 pKa = 10.82IRR28 pKa = 11.84GQLEE32 pKa = 3.78MANNQGADD40 pKa = 2.8TGYY43 pKa = 10.91LHH45 pKa = 6.57WQLLVVFSRR54 pKa = 11.84KK55 pKa = 9.08CRR57 pKa = 11.84LAAVKK62 pKa = 10.15KK63 pKa = 10.15LLGNTVHH70 pKa = 7.26AEE72 pKa = 4.33LSRR75 pKa = 11.84SAAATDD81 pKa = 4.06YY82 pKa = 10.8VWKK85 pKa = 10.25EE86 pKa = 3.95DD87 pKa = 3.4TRR89 pKa = 11.84VAGTQFEE96 pKa = 4.68LGAMPMQRR104 pKa = 11.84NNPTDD109 pKa = 3.26WDD111 pKa = 4.05TVRR114 pKa = 11.84NNARR118 pKa = 11.84AGRR121 pKa = 11.84LGDD124 pKa = 3.39ISGDD128 pKa = 3.04IYY130 pKa = 10.9VRR132 pKa = 11.84CYY134 pKa = 10.79NQLKK138 pKa = 10.45RR139 pKa = 11.84IACDD143 pKa = 3.12HH144 pKa = 5.78MQADD148 pKa = 3.85PVEE151 pKa = 4.98RR152 pKa = 11.84EE153 pKa = 4.32VVCYY157 pKa = 9.39WGRR160 pKa = 11.84TGTGKK165 pKa = 9.99SRR167 pKa = 11.84RR168 pKa = 11.84AWDD171 pKa = 3.64EE172 pKa = 3.86ASLDD176 pKa = 4.31AYY178 pKa = 10.14PKK180 pKa = 10.45DD181 pKa = 3.65PRR183 pKa = 11.84SKK185 pKa = 10.25FWDD188 pKa = 4.41GYY190 pKa = 9.92RR191 pKa = 11.84GQEE194 pKa = 3.8HH195 pKa = 5.89VVLDD199 pKa = 4.06EE200 pKa = 4.0FRR202 pKa = 11.84GGIDD206 pKa = 2.92IAHH209 pKa = 6.28VLRR212 pKa = 11.84WFDD215 pKa = 3.75RR216 pKa = 11.84YY217 pKa = 10.29PVMVEE222 pKa = 3.84VKK224 pKa = 10.41GSSVPLKK231 pKa = 9.61ATKK234 pKa = 9.7IWITSNLPPEE244 pKa = 4.05SWYY247 pKa = 10.32PEE249 pKa = 4.13LDD251 pKa = 4.1DD252 pKa = 3.99EE253 pKa = 4.86TKK255 pKa = 9.75QALRR259 pKa = 11.84RR260 pKa = 11.84RR261 pKa = 11.84MNITHH266 pKa = 7.51FII268 pKa = 3.93
MM1 pKa = 7.16SQGIYY6 pKa = 9.63WLLTINQHH14 pKa = 5.28EE15 pKa = 4.51FVPYY19 pKa = 10.13LPAGVSYY26 pKa = 10.82IRR28 pKa = 11.84GQLEE32 pKa = 3.78MANNQGADD40 pKa = 2.8TGYY43 pKa = 10.91LHH45 pKa = 6.57WQLLVVFSRR54 pKa = 11.84KK55 pKa = 9.08CRR57 pKa = 11.84LAAVKK62 pKa = 10.15KK63 pKa = 10.15LLGNTVHH70 pKa = 7.26AEE72 pKa = 4.33LSRR75 pKa = 11.84SAAATDD81 pKa = 4.06YY82 pKa = 10.8VWKK85 pKa = 10.25EE86 pKa = 3.95DD87 pKa = 3.4TRR89 pKa = 11.84VAGTQFEE96 pKa = 4.68LGAMPMQRR104 pKa = 11.84NNPTDD109 pKa = 3.26WDD111 pKa = 4.05TVRR114 pKa = 11.84NNARR118 pKa = 11.84AGRR121 pKa = 11.84LGDD124 pKa = 3.39ISGDD128 pKa = 3.04IYY130 pKa = 10.9VRR132 pKa = 11.84CYY134 pKa = 10.79NQLKK138 pKa = 10.45RR139 pKa = 11.84IACDD143 pKa = 3.12HH144 pKa = 5.78MQADD148 pKa = 3.85PVEE151 pKa = 4.98RR152 pKa = 11.84EE153 pKa = 4.32VVCYY157 pKa = 9.39WGRR160 pKa = 11.84TGTGKK165 pKa = 9.99SRR167 pKa = 11.84RR168 pKa = 11.84AWDD171 pKa = 3.64EE172 pKa = 3.86ASLDD176 pKa = 4.31AYY178 pKa = 10.14PKK180 pKa = 10.45DD181 pKa = 3.65PRR183 pKa = 11.84SKK185 pKa = 10.25FWDD188 pKa = 4.41GYY190 pKa = 9.92RR191 pKa = 11.84GQEE194 pKa = 3.8HH195 pKa = 5.89VVLDD199 pKa = 4.06EE200 pKa = 4.0FRR202 pKa = 11.84GGIDD206 pKa = 2.92IAHH209 pKa = 6.28VLRR212 pKa = 11.84WFDD215 pKa = 3.75RR216 pKa = 11.84YY217 pKa = 10.29PVMVEE222 pKa = 3.84VKK224 pKa = 10.41GSSVPLKK231 pKa = 9.61ATKK234 pKa = 9.7IWITSNLPPEE244 pKa = 4.05SWYY247 pKa = 10.32PEE249 pKa = 4.13LDD251 pKa = 4.1DD252 pKa = 3.99EE253 pKa = 4.86TKK255 pKa = 9.75QALRR259 pKa = 11.84RR260 pKa = 11.84RR261 pKa = 11.84MNITHH266 pKa = 7.51FII268 pKa = 3.93
Molecular weight: 30.84 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140AQJ3|A0A140AQJ3_9VIRU Replication associated protein OS=Lake Sarah-associated circular virus-1 OX=1685735 PE=4 SV=1
MM1 pKa = 7.18KK2 pKa = 10.33SKK4 pKa = 10.45YY5 pKa = 9.59VKK7 pKa = 10.11LAARR11 pKa = 11.84VAPYY15 pKa = 10.36AAIAGRR21 pKa = 11.84QLVRR25 pKa = 11.84YY26 pKa = 8.89AGNRR30 pKa = 11.84AKK32 pKa = 10.69DD33 pKa = 3.63YY34 pKa = 11.2VYY36 pKa = 10.74GKK38 pKa = 10.28SSSKK42 pKa = 9.5PKK44 pKa = 10.64SRR46 pKa = 11.84ISPPKK51 pKa = 9.58AKK53 pKa = 10.71ANFGFRR59 pKa = 11.84GTGKK63 pKa = 10.52LGGFVGRR70 pKa = 11.84GKK72 pKa = 10.34RR73 pKa = 11.84VRR75 pKa = 11.84KK76 pKa = 8.06TKK78 pKa = 10.42FSQAQRR84 pKa = 11.84YY85 pKa = 8.07GVYY88 pKa = 9.71QQVEE92 pKa = 4.41IYY94 pKa = 10.56KK95 pKa = 10.31SVTGSQTLAVGHH107 pKa = 6.21ATNPVKK113 pKa = 10.9VVFGSVAHH121 pKa = 6.68ALLKK125 pKa = 10.89KK126 pKa = 10.27FVEE129 pKa = 4.19RR130 pKa = 11.84DD131 pKa = 3.79AICFSDD137 pKa = 3.31WNTVVVGNTKK147 pKa = 10.37YY148 pKa = 8.32PTGAKK153 pKa = 10.08FYY155 pKa = 10.48ISYY158 pKa = 10.43RR159 pKa = 11.84DD160 pKa = 3.31TDD162 pKa = 3.91AGTMADD168 pKa = 3.97SLVITIGGATTFEE181 pKa = 5.28DD182 pKa = 3.21IALAIYY188 pKa = 7.41TTISTLSSKK197 pKa = 11.16YY198 pKa = 11.03SMQLMYY204 pKa = 10.89AVTGAGLLLTTLYY217 pKa = 11.09LNTAFVKK224 pKa = 10.56LHH226 pKa = 6.1TKK228 pKa = 10.23SALKK232 pKa = 9.76FQNITIDD239 pKa = 3.39TGASTEE245 pKa = 3.92QDD247 pKa = 3.12DD248 pKa = 5.57VRR250 pKa = 11.84NQPLNGKK257 pKa = 9.46AYY259 pKa = 9.1EE260 pKa = 4.44GKK262 pKa = 8.75GTGADD267 pKa = 3.95VYY269 pKa = 10.6WIGAGAASNLVCDD282 pKa = 3.58TAYY285 pKa = 10.67GVLGVSDD292 pKa = 4.33PVEE295 pKa = 5.68DD296 pKa = 3.54IPQPKK301 pKa = 9.16VVQRR305 pKa = 11.84ASKK308 pKa = 9.69FSKK311 pKa = 10.32VRR313 pKa = 11.84IDD315 pKa = 3.16AGAIKK320 pKa = 9.9TSVLTTNHH328 pKa = 7.66KK329 pKa = 10.27IRR331 pKa = 11.84MSDD334 pKa = 3.49LVSMLNSGVEE344 pKa = 4.4GYY346 pKa = 11.16NRR348 pKa = 11.84MGKK351 pKa = 9.52FRR353 pKa = 11.84IFILEE358 pKa = 3.88RR359 pKa = 11.84LIGGNFGSATMQVRR373 pKa = 11.84IEE375 pKa = 4.17HH376 pKa = 6.18QLDD379 pKa = 3.28QSATVIGGNPPYY391 pKa = 10.2MKK393 pKa = 9.68TKK395 pKa = 10.4FVAVQLL401 pKa = 4.08
MM1 pKa = 7.18KK2 pKa = 10.33SKK4 pKa = 10.45YY5 pKa = 9.59VKK7 pKa = 10.11LAARR11 pKa = 11.84VAPYY15 pKa = 10.36AAIAGRR21 pKa = 11.84QLVRR25 pKa = 11.84YY26 pKa = 8.89AGNRR30 pKa = 11.84AKK32 pKa = 10.69DD33 pKa = 3.63YY34 pKa = 11.2VYY36 pKa = 10.74GKK38 pKa = 10.28SSSKK42 pKa = 9.5PKK44 pKa = 10.64SRR46 pKa = 11.84ISPPKK51 pKa = 9.58AKK53 pKa = 10.71ANFGFRR59 pKa = 11.84GTGKK63 pKa = 10.52LGGFVGRR70 pKa = 11.84GKK72 pKa = 10.34RR73 pKa = 11.84VRR75 pKa = 11.84KK76 pKa = 8.06TKK78 pKa = 10.42FSQAQRR84 pKa = 11.84YY85 pKa = 8.07GVYY88 pKa = 9.71QQVEE92 pKa = 4.41IYY94 pKa = 10.56KK95 pKa = 10.31SVTGSQTLAVGHH107 pKa = 6.21ATNPVKK113 pKa = 10.9VVFGSVAHH121 pKa = 6.68ALLKK125 pKa = 10.89KK126 pKa = 10.27FVEE129 pKa = 4.19RR130 pKa = 11.84DD131 pKa = 3.79AICFSDD137 pKa = 3.31WNTVVVGNTKK147 pKa = 10.37YY148 pKa = 8.32PTGAKK153 pKa = 10.08FYY155 pKa = 10.48ISYY158 pKa = 10.43RR159 pKa = 11.84DD160 pKa = 3.31TDD162 pKa = 3.91AGTMADD168 pKa = 3.97SLVITIGGATTFEE181 pKa = 5.28DD182 pKa = 3.21IALAIYY188 pKa = 7.41TTISTLSSKK197 pKa = 11.16YY198 pKa = 11.03SMQLMYY204 pKa = 10.89AVTGAGLLLTTLYY217 pKa = 11.09LNTAFVKK224 pKa = 10.56LHH226 pKa = 6.1TKK228 pKa = 10.23SALKK232 pKa = 9.76FQNITIDD239 pKa = 3.39TGASTEE245 pKa = 3.92QDD247 pKa = 3.12DD248 pKa = 5.57VRR250 pKa = 11.84NQPLNGKK257 pKa = 9.46AYY259 pKa = 9.1EE260 pKa = 4.44GKK262 pKa = 8.75GTGADD267 pKa = 3.95VYY269 pKa = 10.6WIGAGAASNLVCDD282 pKa = 3.58TAYY285 pKa = 10.67GVLGVSDD292 pKa = 4.33PVEE295 pKa = 5.68DD296 pKa = 3.54IPQPKK301 pKa = 9.16VVQRR305 pKa = 11.84ASKK308 pKa = 9.69FSKK311 pKa = 10.32VRR313 pKa = 11.84IDD315 pKa = 3.16AGAIKK320 pKa = 9.9TSVLTTNHH328 pKa = 7.66KK329 pKa = 10.27IRR331 pKa = 11.84MSDD334 pKa = 3.49LVSMLNSGVEE344 pKa = 4.4GYY346 pKa = 11.16NRR348 pKa = 11.84MGKK351 pKa = 9.52FRR353 pKa = 11.84IFILEE358 pKa = 3.88RR359 pKa = 11.84LIGGNFGSATMQVRR373 pKa = 11.84IEE375 pKa = 4.17HH376 pKa = 6.18QLDD379 pKa = 3.28QSATVIGGNPPYY391 pKa = 10.2MKK393 pKa = 9.68TKK395 pKa = 10.4FVAVQLL401 pKa = 4.08
Molecular weight: 43.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
669 |
268 |
401 |
334.5 |
37.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.969 ± 0.699 | 0.897 ± 0.367 |
5.232 ± 0.916 | 3.438 ± 1.101 |
3.438 ± 0.509 | 8.67 ± 0.975 |
1.794 ± 0.505 | 5.082 ± 0.373 |
6.726 ± 1.387 | 7.175 ± 0.408 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.392 ± 0.136 | 3.886 ± 0.135 |
3.587 ± 0.549 | 4.036 ± 0.042 |
6.428 ± 1.329 | 6.278 ± 0.88 |
6.876 ± 1.019 | 8.52 ± 0.652 |
1.794 ± 1.195 | 4.783 ± 0.189 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
