
Modestobacter versicolor
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Geodermatophilales; Geodermatophilaceae; Modestobacter
Average proteome isoelectric point is 6.03
Get precalculated fractions of proteins
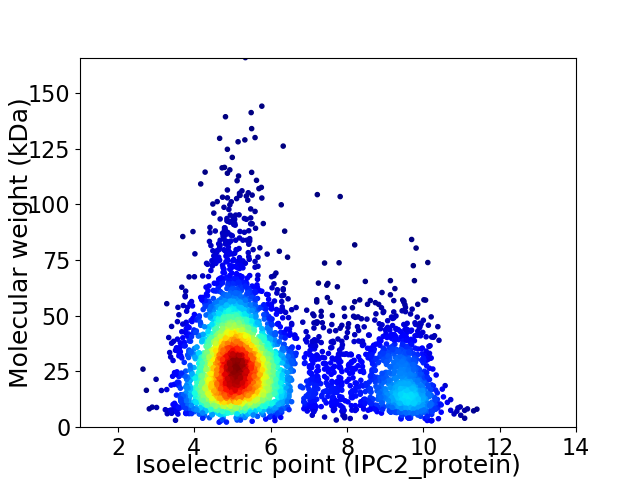
Virtual 2D-PAGE plot for 4717 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A323VIT2|A0A323VIT2_9ACTN ATP-dependent DNA ligase OS=Modestobacter versicolor OX=429133 GN=DMO24_02595 PE=4 SV=1
MM1 pKa = 7.44KK2 pKa = 10.64LSTTTRR8 pKa = 11.84AAAVSASLAAVLALAACGASNEE30 pKa = 4.52DD31 pKa = 3.64SGSDD35 pKa = 3.33SGSGGGSDD43 pKa = 4.13SAAEE47 pKa = 4.0LSGSLVGAGASSQQSAMEE65 pKa = 4.09AWQAGFEE72 pKa = 4.25GEE74 pKa = 4.28YY75 pKa = 10.32PDD77 pKa = 4.36VQFSYY82 pKa = 11.11APEE85 pKa = 4.41GSGAGRR91 pKa = 11.84EE92 pKa = 3.98QFINGSADD100 pKa = 3.34FAGSDD105 pKa = 3.21AALDD109 pKa = 4.03EE110 pKa = 4.83EE111 pKa = 4.79EE112 pKa = 4.35LTAASEE118 pKa = 4.26RR119 pKa = 11.84CAGGEE124 pKa = 4.12VFEE127 pKa = 4.74LPNYY131 pKa = 9.76ISPIAVAFNLEE142 pKa = 4.38GVDD145 pKa = 4.06EE146 pKa = 5.03LNLTPATIANIFNGTITTWNDD167 pKa = 2.9PAIAADD173 pKa = 3.83NADD176 pKa = 3.42ATLPATAITPVHH188 pKa = 6.42RR189 pKa = 11.84ADD191 pKa = 5.34DD192 pKa = 3.86SGTTEE197 pKa = 3.92NFTDD201 pKa = 4.15YY202 pKa = 10.94LHH204 pKa = 6.51QAAADD209 pKa = 3.52VWTAEE214 pKa = 4.25PDD216 pKa = 4.15GVWPLEE222 pKa = 4.37GGEE225 pKa = 4.36SAQGTSGVIEE235 pKa = 4.25VVTNTVGAITYY246 pKa = 10.35ADD248 pKa = 3.63ASRR251 pKa = 11.84AGDD254 pKa = 3.48LGVANVAVGSDD265 pKa = 4.03FVAPSPEE272 pKa = 3.37AAAAVVEE279 pKa = 4.11NSPAVEE285 pKa = 4.06GRR287 pKa = 11.84GEE289 pKa = 3.91YY290 pKa = 10.87DD291 pKa = 3.38FAIEE295 pKa = 4.24VNRR298 pKa = 11.84LTEE301 pKa = 4.05GTGEE305 pKa = 4.2YY306 pKa = 10.05PVVLVSYY313 pKa = 10.52HH314 pKa = 6.26IGCVEE319 pKa = 4.0YY320 pKa = 10.69DD321 pKa = 3.86DD322 pKa = 5.42QEE324 pKa = 4.23KK325 pKa = 10.99ADD327 pKa = 3.8NVKK330 pKa = 11.06AFMNYY335 pKa = 9.44VISEE339 pKa = 4.37DD340 pKa = 3.85GQATAAEE347 pKa = 4.34NAGNAQISDD356 pKa = 3.95TLRR359 pKa = 11.84EE360 pKa = 4.06QAQGAVDD367 pKa = 5.53AITAAGG373 pKa = 3.34
MM1 pKa = 7.44KK2 pKa = 10.64LSTTTRR8 pKa = 11.84AAAVSASLAAVLALAACGASNEE30 pKa = 4.52DD31 pKa = 3.64SGSDD35 pKa = 3.33SGSGGGSDD43 pKa = 4.13SAAEE47 pKa = 4.0LSGSLVGAGASSQQSAMEE65 pKa = 4.09AWQAGFEE72 pKa = 4.25GEE74 pKa = 4.28YY75 pKa = 10.32PDD77 pKa = 4.36VQFSYY82 pKa = 11.11APEE85 pKa = 4.41GSGAGRR91 pKa = 11.84EE92 pKa = 3.98QFINGSADD100 pKa = 3.34FAGSDD105 pKa = 3.21AALDD109 pKa = 4.03EE110 pKa = 4.83EE111 pKa = 4.79EE112 pKa = 4.35LTAASEE118 pKa = 4.26RR119 pKa = 11.84CAGGEE124 pKa = 4.12VFEE127 pKa = 4.74LPNYY131 pKa = 9.76ISPIAVAFNLEE142 pKa = 4.38GVDD145 pKa = 4.06EE146 pKa = 5.03LNLTPATIANIFNGTITTWNDD167 pKa = 2.9PAIAADD173 pKa = 3.83NADD176 pKa = 3.42ATLPATAITPVHH188 pKa = 6.42RR189 pKa = 11.84ADD191 pKa = 5.34DD192 pKa = 3.86SGTTEE197 pKa = 3.92NFTDD201 pKa = 4.15YY202 pKa = 10.94LHH204 pKa = 6.51QAAADD209 pKa = 3.52VWTAEE214 pKa = 4.25PDD216 pKa = 4.15GVWPLEE222 pKa = 4.37GGEE225 pKa = 4.36SAQGTSGVIEE235 pKa = 4.25VVTNTVGAITYY246 pKa = 10.35ADD248 pKa = 3.63ASRR251 pKa = 11.84AGDD254 pKa = 3.48LGVANVAVGSDD265 pKa = 4.03FVAPSPEE272 pKa = 3.37AAAAVVEE279 pKa = 4.11NSPAVEE285 pKa = 4.06GRR287 pKa = 11.84GEE289 pKa = 3.91YY290 pKa = 10.87DD291 pKa = 3.38FAIEE295 pKa = 4.24VNRR298 pKa = 11.84LTEE301 pKa = 4.05GTGEE305 pKa = 4.2YY306 pKa = 10.05PVVLVSYY313 pKa = 10.52HH314 pKa = 6.26IGCVEE319 pKa = 4.0YY320 pKa = 10.69DD321 pKa = 3.86DD322 pKa = 5.42QEE324 pKa = 4.23KK325 pKa = 10.99ADD327 pKa = 3.8NVKK330 pKa = 11.06AFMNYY335 pKa = 9.44VISEE339 pKa = 4.37DD340 pKa = 3.85GQATAAEE347 pKa = 4.34NAGNAQISDD356 pKa = 3.95TLRR359 pKa = 11.84EE360 pKa = 4.06QAQGAVDD367 pKa = 5.53AITAAGG373 pKa = 3.34
Molecular weight: 37.84 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A323V9V3|A0A323V9V3_9ACTN Uncharacterized protein OS=Modestobacter versicolor OX=429133 GN=DMO24_13190 PE=4 SV=1
MM1 pKa = 7.56SILSKK6 pKa = 11.12LMGTAQQTAGRR17 pKa = 11.84GRR19 pKa = 11.84RR20 pKa = 11.84VGGRR24 pKa = 11.84PATGRR29 pKa = 11.84RR30 pKa = 11.84TTGTGRR36 pKa = 11.84TAGGTARR43 pKa = 11.84MARR46 pKa = 11.84GTTSRR51 pKa = 11.84GRR53 pKa = 11.84AGAPAAGGLGKK64 pKa = 10.47LLSSFAKK71 pKa = 10.16RR72 pKa = 11.84RR73 pKa = 3.58
MM1 pKa = 7.56SILSKK6 pKa = 11.12LMGTAQQTAGRR17 pKa = 11.84GRR19 pKa = 11.84RR20 pKa = 11.84VGGRR24 pKa = 11.84PATGRR29 pKa = 11.84RR30 pKa = 11.84TTGTGRR36 pKa = 11.84TAGGTARR43 pKa = 11.84MARR46 pKa = 11.84GTTSRR51 pKa = 11.84GRR53 pKa = 11.84AGAPAAGGLGKK64 pKa = 10.47LLSSFAKK71 pKa = 10.16RR72 pKa = 11.84RR73 pKa = 3.58
Molecular weight: 7.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1332854 |
24 |
1538 |
282.6 |
29.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.34 ± 0.05 | 0.65 ± 0.01 |
6.233 ± 0.031 | 5.442 ± 0.034 |
2.596 ± 0.024 | 9.663 ± 0.03 |
2.001 ± 0.02 | 2.698 ± 0.03 |
1.353 ± 0.025 | 10.806 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.578 ± 0.013 | 1.43 ± 0.016 |
6.154 ± 0.026 | 2.921 ± 0.021 |
7.75 ± 0.039 | 4.916 ± 0.024 |
6.197 ± 0.029 | 10.093 ± 0.04 |
1.459 ± 0.016 | 1.72 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
