
Palaemonetes intermedius brackish grass shrimp associated circular virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 8.19
Get precalculated fractions of proteins
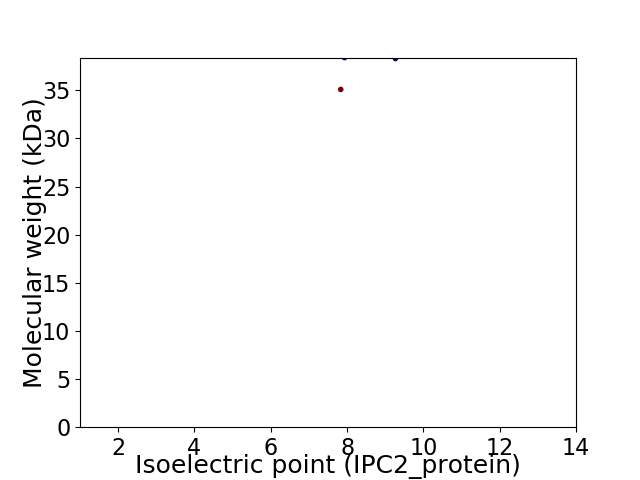
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K1RL48|A0A0K1RL48_9CIRC Putative capsid protein OS=Palaemonetes intermedius brackish grass shrimp associated circular virus OX=1692257 PE=4 SV=1
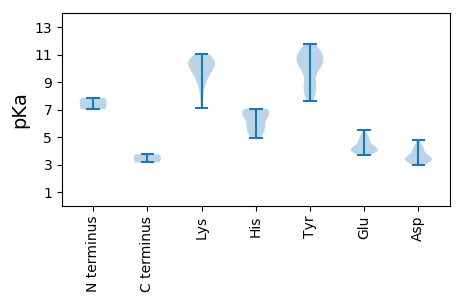
MM1 pKa = 7.85NIRR4 pKa = 11.84PAAVKK9 pKa = 10.14RR10 pKa = 11.84WVATVNNPTQQEE22 pKa = 4.46SQTLRR27 pKa = 11.84DD28 pKa = 4.08AIEE31 pKa = 4.24AQTSYY36 pKa = 11.44AVIGRR41 pKa = 11.84EE42 pKa = 4.09VGEE45 pKa = 4.37SGTPHH50 pKa = 6.6LQCFFIFNNRR60 pKa = 11.84LRR62 pKa = 11.84LRR64 pKa = 11.84QVKK67 pKa = 9.48AVPGLQRR74 pKa = 11.84AHH76 pKa = 7.06LEE78 pKa = 4.03PARR81 pKa = 11.84GTSAQAANYY90 pKa = 8.03CKK92 pKa = 10.15KK93 pKa = 10.76DD94 pKa = 3.21GDD96 pKa = 3.55FDD98 pKa = 4.57EE99 pKa = 5.52YY100 pKa = 11.78GEE102 pKa = 4.87LPNSGPKK109 pKa = 7.81TTIFEE114 pKa = 4.81AFRR117 pKa = 11.84DD118 pKa = 3.88WYY120 pKa = 10.38KK121 pKa = 10.82DD122 pKa = 3.22QPGVVTEE129 pKa = 4.22RR130 pKa = 11.84DD131 pKa = 3.5ILDD134 pKa = 3.45HH135 pKa = 6.69HH136 pKa = 6.83PSILRR141 pKa = 11.84YY142 pKa = 7.61PHH144 pKa = 6.86FIEE147 pKa = 4.52VCHH150 pKa = 5.76RR151 pKa = 11.84QYY153 pKa = 11.43GRR155 pKa = 11.84RR156 pKa = 11.84PTLVEE161 pKa = 3.76GQLRR165 pKa = 11.84NWQLEE170 pKa = 4.24LSNMLDD176 pKa = 3.62EE177 pKa = 5.22EE178 pKa = 5.02PDD180 pKa = 3.37DD181 pKa = 4.77RR182 pKa = 11.84KK183 pKa = 10.48ICFVVDD189 pKa = 3.24EE190 pKa = 5.05EE191 pKa = 4.76GNKK194 pKa = 9.51GKK196 pKa = 10.19SWLTAYY202 pKa = 9.03WYY204 pKa = 10.83SNRR207 pKa = 11.84SDD209 pKa = 3.43VQMLSIGKK217 pKa = 9.51RR218 pKa = 11.84DD219 pKa = 3.95DD220 pKa = 3.06LTYY223 pKa = 10.75AIDD226 pKa = 3.34VSKK229 pKa = 10.75RR230 pKa = 11.84VFVFDD235 pKa = 3.63INRR238 pKa = 11.84GQMEE242 pKa = 4.4YY243 pKa = 10.24FQYY246 pKa = 11.22SVVEE250 pKa = 4.06SLKK253 pKa = 10.81NRR255 pKa = 11.84MIMSNKK261 pKa = 8.66YY262 pKa = 10.2KK263 pKa = 10.47SVTKK267 pKa = 10.45IIPHH271 pKa = 5.99KK272 pKa = 8.39VHH274 pKa = 6.8VIVFCNEE281 pKa = 3.73EE282 pKa = 3.97PDD284 pKa = 3.58RR285 pKa = 11.84TAMTRR290 pKa = 11.84DD291 pKa = 3.32RR292 pKa = 11.84YY293 pKa = 8.68QMKK296 pKa = 10.54RR297 pKa = 11.84ITPLL301 pKa = 3.16
MM1 pKa = 7.85NIRR4 pKa = 11.84PAAVKK9 pKa = 10.14RR10 pKa = 11.84WVATVNNPTQQEE22 pKa = 4.46SQTLRR27 pKa = 11.84DD28 pKa = 4.08AIEE31 pKa = 4.24AQTSYY36 pKa = 11.44AVIGRR41 pKa = 11.84EE42 pKa = 4.09VGEE45 pKa = 4.37SGTPHH50 pKa = 6.6LQCFFIFNNRR60 pKa = 11.84LRR62 pKa = 11.84LRR64 pKa = 11.84QVKK67 pKa = 9.48AVPGLQRR74 pKa = 11.84AHH76 pKa = 7.06LEE78 pKa = 4.03PARR81 pKa = 11.84GTSAQAANYY90 pKa = 8.03CKK92 pKa = 10.15KK93 pKa = 10.76DD94 pKa = 3.21GDD96 pKa = 3.55FDD98 pKa = 4.57EE99 pKa = 5.52YY100 pKa = 11.78GEE102 pKa = 4.87LPNSGPKK109 pKa = 7.81TTIFEE114 pKa = 4.81AFRR117 pKa = 11.84DD118 pKa = 3.88WYY120 pKa = 10.38KK121 pKa = 10.82DD122 pKa = 3.22QPGVVTEE129 pKa = 4.22RR130 pKa = 11.84DD131 pKa = 3.5ILDD134 pKa = 3.45HH135 pKa = 6.69HH136 pKa = 6.83PSILRR141 pKa = 11.84YY142 pKa = 7.61PHH144 pKa = 6.86FIEE147 pKa = 4.52VCHH150 pKa = 5.76RR151 pKa = 11.84QYY153 pKa = 11.43GRR155 pKa = 11.84RR156 pKa = 11.84PTLVEE161 pKa = 3.76GQLRR165 pKa = 11.84NWQLEE170 pKa = 4.24LSNMLDD176 pKa = 3.62EE177 pKa = 5.22EE178 pKa = 5.02PDD180 pKa = 3.37DD181 pKa = 4.77RR182 pKa = 11.84KK183 pKa = 10.48ICFVVDD189 pKa = 3.24EE190 pKa = 5.05EE191 pKa = 4.76GNKK194 pKa = 9.51GKK196 pKa = 10.19SWLTAYY202 pKa = 9.03WYY204 pKa = 10.83SNRR207 pKa = 11.84SDD209 pKa = 3.43VQMLSIGKK217 pKa = 9.51RR218 pKa = 11.84DD219 pKa = 3.95DD220 pKa = 3.06LTYY223 pKa = 10.75AIDD226 pKa = 3.34VSKK229 pKa = 10.75RR230 pKa = 11.84VFVFDD235 pKa = 3.63INRR238 pKa = 11.84GQMEE242 pKa = 4.4YY243 pKa = 10.24FQYY246 pKa = 11.22SVVEE250 pKa = 4.06SLKK253 pKa = 10.81NRR255 pKa = 11.84MIMSNKK261 pKa = 8.66YY262 pKa = 10.2KK263 pKa = 10.47SVTKK267 pKa = 10.45IIPHH271 pKa = 5.99KK272 pKa = 8.39VHH274 pKa = 6.8VIVFCNEE281 pKa = 3.73EE282 pKa = 3.97PDD284 pKa = 3.58RR285 pKa = 11.84TAMTRR290 pKa = 11.84DD291 pKa = 3.32RR292 pKa = 11.84YY293 pKa = 8.68QMKK296 pKa = 10.54RR297 pKa = 11.84ITPLL301 pKa = 3.16
Molecular weight: 35.08 kDa
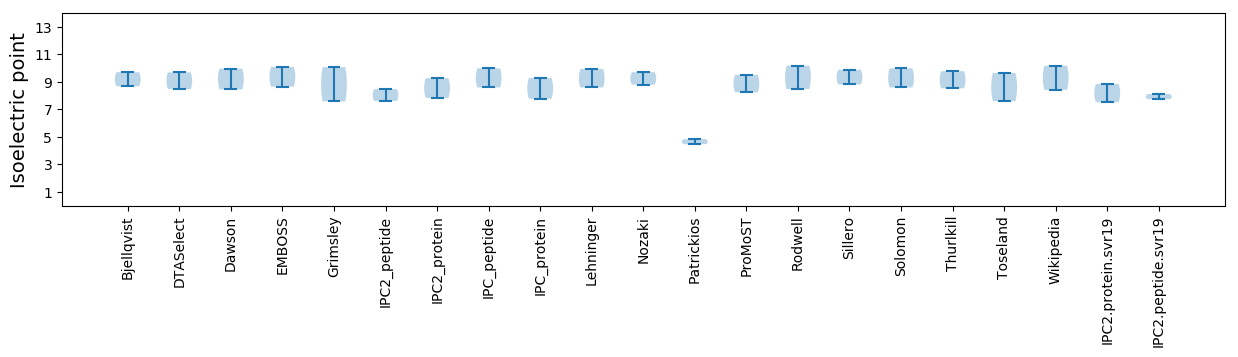
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K1RL48|A0A0K1RL48_9CIRC Putative capsid protein OS=Palaemonetes intermedius brackish grass shrimp associated circular virus OX=1692257 PE=4 SV=1
MM1 pKa = 7.03QAGLAALAEE10 pKa = 4.25GDD12 pKa = 4.17YY13 pKa = 10.35IYY15 pKa = 10.74PLAQYY20 pKa = 11.07AGLEE24 pKa = 4.17LLASAAARR32 pKa = 11.84HH33 pKa = 5.49GSNRR37 pKa = 11.84GTYY40 pKa = 8.5MPAEE44 pKa = 3.96SFLRR48 pKa = 11.84KK49 pKa = 9.17RR50 pKa = 11.84AAVTPDD56 pKa = 2.96EE57 pKa = 4.78PQTKK61 pKa = 9.49RR62 pKa = 11.84QRR64 pKa = 11.84TYY66 pKa = 8.8TYY68 pKa = 10.68APRR71 pKa = 11.84VLNMPAGVMTRR82 pKa = 11.84TRR84 pKa = 11.84RR85 pKa = 11.84TVYY88 pKa = 10.04KK89 pKa = 9.84YY90 pKa = 11.07NLGTRR95 pKa = 11.84VGKK98 pKa = 8.88FKK100 pKa = 11.05CNKK103 pKa = 9.05FSFKK107 pKa = 10.56TGVTNNMADD116 pKa = 3.53KK117 pKa = 10.22AKK119 pKa = 10.61HH120 pKa = 4.9SFRR123 pKa = 11.84LIQIPYY129 pKa = 10.18SSDD132 pKa = 3.15DD133 pKa = 3.4SLFNTRR139 pKa = 11.84NGRR142 pKa = 11.84LVNVRR147 pKa = 11.84GVRR150 pKa = 11.84LQWVCEE156 pKa = 3.97LKK158 pKa = 10.84PGGSTGNIEE167 pKa = 4.2RR168 pKa = 11.84PLSVRR173 pKa = 11.84WAIINPKK180 pKa = 9.74EE181 pKa = 3.93NDD183 pKa = 3.48GTSTVGTANWWTSLNPTNEE202 pKa = 3.94YY203 pKa = 9.67ATDD206 pKa = 4.31FVNGSGSTTPGTGTGYY222 pKa = 10.71QDD224 pKa = 3.28WFDD227 pKa = 4.04LMTRR231 pKa = 11.84GINRR235 pKa = 11.84RR236 pKa = 11.84KK237 pKa = 10.35YY238 pKa = 8.67GVLKK242 pKa = 10.14EE243 pKa = 4.0GHH245 pKa = 5.42FTISPPEE252 pKa = 3.94TSDD255 pKa = 3.13ADD257 pKa = 3.67SRR259 pKa = 11.84KK260 pKa = 7.12TWRR263 pKa = 11.84QQKK266 pKa = 10.06VIQTYY271 pKa = 10.71LKK273 pKa = 10.37INQCMKK279 pKa = 9.69WGSNNTGGEE288 pKa = 3.86NDD290 pKa = 3.71YY291 pKa = 11.34PNANLYY297 pKa = 8.01FVWWYY302 pKa = 10.63CDD304 pKa = 3.23RR305 pKa = 11.84GNMDD309 pKa = 4.3DD310 pKa = 4.19LQWYY314 pKa = 8.47PSVNYY319 pKa = 10.07VPLQTQAQVTVFFKK333 pKa = 10.02TAKK336 pKa = 10.43GLLL339 pKa = 3.75
MM1 pKa = 7.03QAGLAALAEE10 pKa = 4.25GDD12 pKa = 4.17YY13 pKa = 10.35IYY15 pKa = 10.74PLAQYY20 pKa = 11.07AGLEE24 pKa = 4.17LLASAAARR32 pKa = 11.84HH33 pKa = 5.49GSNRR37 pKa = 11.84GTYY40 pKa = 8.5MPAEE44 pKa = 3.96SFLRR48 pKa = 11.84KK49 pKa = 9.17RR50 pKa = 11.84AAVTPDD56 pKa = 2.96EE57 pKa = 4.78PQTKK61 pKa = 9.49RR62 pKa = 11.84QRR64 pKa = 11.84TYY66 pKa = 8.8TYY68 pKa = 10.68APRR71 pKa = 11.84VLNMPAGVMTRR82 pKa = 11.84TRR84 pKa = 11.84RR85 pKa = 11.84TVYY88 pKa = 10.04KK89 pKa = 9.84YY90 pKa = 11.07NLGTRR95 pKa = 11.84VGKK98 pKa = 8.88FKK100 pKa = 11.05CNKK103 pKa = 9.05FSFKK107 pKa = 10.56TGVTNNMADD116 pKa = 3.53KK117 pKa = 10.22AKK119 pKa = 10.61HH120 pKa = 4.9SFRR123 pKa = 11.84LIQIPYY129 pKa = 10.18SSDD132 pKa = 3.15DD133 pKa = 3.4SLFNTRR139 pKa = 11.84NGRR142 pKa = 11.84LVNVRR147 pKa = 11.84GVRR150 pKa = 11.84LQWVCEE156 pKa = 3.97LKK158 pKa = 10.84PGGSTGNIEE167 pKa = 4.2RR168 pKa = 11.84PLSVRR173 pKa = 11.84WAIINPKK180 pKa = 9.74EE181 pKa = 3.93NDD183 pKa = 3.48GTSTVGTANWWTSLNPTNEE202 pKa = 3.94YY203 pKa = 9.67ATDD206 pKa = 4.31FVNGSGSTTPGTGTGYY222 pKa = 10.71QDD224 pKa = 3.28WFDD227 pKa = 4.04LMTRR231 pKa = 11.84GINRR235 pKa = 11.84RR236 pKa = 11.84KK237 pKa = 10.35YY238 pKa = 8.67GVLKK242 pKa = 10.14EE243 pKa = 4.0GHH245 pKa = 5.42FTISPPEE252 pKa = 3.94TSDD255 pKa = 3.13ADD257 pKa = 3.67SRR259 pKa = 11.84KK260 pKa = 7.12TWRR263 pKa = 11.84QQKK266 pKa = 10.06VIQTYY271 pKa = 10.71LKK273 pKa = 10.37INQCMKK279 pKa = 9.69WGSNNTGGEE288 pKa = 3.86NDD290 pKa = 3.71YY291 pKa = 11.34PNANLYY297 pKa = 8.01FVWWYY302 pKa = 10.63CDD304 pKa = 3.23RR305 pKa = 11.84GNMDD309 pKa = 4.3DD310 pKa = 4.19LQWYY314 pKa = 8.47PSVNYY319 pKa = 10.07VPLQTQAQVTVFFKK333 pKa = 10.02TAKK336 pKa = 10.43GLLL339 pKa = 3.75
Molecular weight: 38.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
640 |
301 |
339 |
320.0 |
36.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.25 ± 0.63 | 1.406 ± 0.172 |
5.313 ± 0.674 | 4.844 ± 1.214 |
3.75 ± 0.16 | 6.875 ± 1.275 |
1.719 ± 0.633 | 4.219 ± 0.963 |
5.625 ± 0.015 | 6.719 ± 0.274 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.5 ± 0.106 | 6.25 ± 0.854 |
5.0 ± 0.011 | 4.844 ± 0.318 |
7.813 ± 0.556 | 5.469 ± 0.103 |
7.5 ± 1.473 | 6.719 ± 0.622 |
2.344 ± 0.46 | 4.844 ± 0.354 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
