
Streptococcus satellite phage Javan285
Taxonomy: Viruses; unclassified bacterial viruses
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
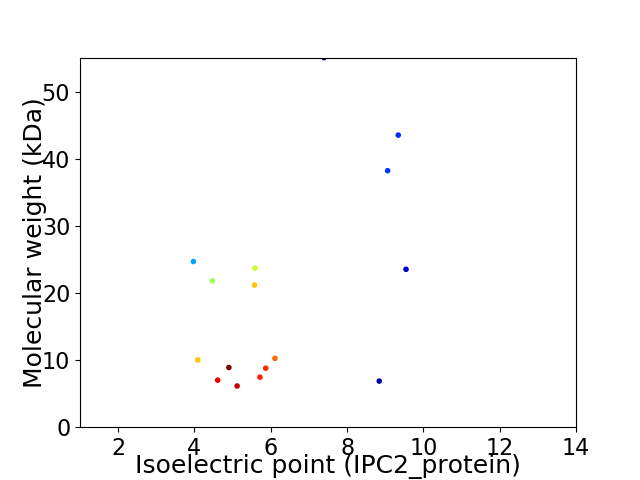
Virtual 2D-PAGE plot for 16 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4D5ZIM1|A0A4D5ZIM1_9VIRU Integrase/recombinase OS=Streptococcus satellite phage Javan285 OX=2558607 GN=JavanS285_0001 PE=3 SV=1
MM1 pKa = 7.61TNTNKK6 pKa = 7.94TTAPKK11 pKa = 9.89TLTEE15 pKa = 4.16LKK17 pKa = 10.37AWTKK21 pKa = 7.97EE22 pKa = 3.45ARR24 pKa = 11.84NWVEE28 pKa = 3.69YY29 pKa = 10.69LQDD32 pKa = 3.71EE33 pKa = 4.74YY34 pKa = 11.06HH35 pKa = 7.11GYY37 pKa = 8.76MPTGHH42 pKa = 6.14QEE44 pKa = 3.99AFGSITNRR52 pKa = 11.84IYY54 pKa = 11.35NCLDD58 pKa = 3.54CLTEE62 pKa = 4.1MFDD65 pKa = 3.92NDD67 pKa = 3.45GLEE70 pKa = 4.1LVEE73 pKa = 4.13YY74 pKa = 9.74TIDD77 pKa = 3.72PQEE80 pKa = 3.98TMIDD84 pKa = 3.57DD85 pKa = 3.88NQANEE90 pKa = 3.79MAQYY94 pKa = 10.55NEE96 pKa = 5.46LMAQYY101 pKa = 10.41NEE103 pKa = 3.98LMKK106 pKa = 10.63EE107 pKa = 4.13VEE109 pKa = 4.64DD110 pKa = 3.69SDD112 pKa = 4.32KK113 pKa = 10.63EE114 pKa = 4.14ARR116 pKa = 11.84EE117 pKa = 3.61IAYY120 pKa = 10.24GDD122 pKa = 4.16TIDD125 pKa = 5.02KK126 pKa = 11.06YY127 pKa = 9.96IDD129 pKa = 3.23KK130 pKa = 10.33RR131 pKa = 11.84AKK133 pKa = 8.97QLKK136 pKa = 8.43EE137 pKa = 3.37QATFDD142 pKa = 3.8EE143 pKa = 5.04LVDD146 pKa = 3.93KK147 pKa = 11.06VALYY151 pKa = 10.43EE152 pKa = 4.89SEE154 pKa = 4.11LLDD157 pKa = 3.57YY158 pKa = 11.28AEE160 pKa = 5.48RR161 pKa = 11.84LLSDD165 pKa = 4.64DD166 pKa = 4.48PLIADD171 pKa = 3.97SEE173 pKa = 4.7TAIGTLEE180 pKa = 3.99MLDD183 pKa = 3.96NEE185 pKa = 5.27AIDD188 pKa = 4.13LFKK191 pKa = 11.29SLDD194 pKa = 3.42IDD196 pKa = 3.65NEE198 pKa = 4.42YY199 pKa = 10.91QGLEE203 pKa = 4.14YY204 pKa = 11.01YY205 pKa = 8.17DD206 pKa = 3.56TSLNKK211 pKa = 9.96EE212 pKa = 4.17DD213 pKa = 3.66
MM1 pKa = 7.61TNTNKK6 pKa = 7.94TTAPKK11 pKa = 9.89TLTEE15 pKa = 4.16LKK17 pKa = 10.37AWTKK21 pKa = 7.97EE22 pKa = 3.45ARR24 pKa = 11.84NWVEE28 pKa = 3.69YY29 pKa = 10.69LQDD32 pKa = 3.71EE33 pKa = 4.74YY34 pKa = 11.06HH35 pKa = 7.11GYY37 pKa = 8.76MPTGHH42 pKa = 6.14QEE44 pKa = 3.99AFGSITNRR52 pKa = 11.84IYY54 pKa = 11.35NCLDD58 pKa = 3.54CLTEE62 pKa = 4.1MFDD65 pKa = 3.92NDD67 pKa = 3.45GLEE70 pKa = 4.1LVEE73 pKa = 4.13YY74 pKa = 9.74TIDD77 pKa = 3.72PQEE80 pKa = 3.98TMIDD84 pKa = 3.57DD85 pKa = 3.88NQANEE90 pKa = 3.79MAQYY94 pKa = 10.55NEE96 pKa = 5.46LMAQYY101 pKa = 10.41NEE103 pKa = 3.98LMKK106 pKa = 10.63EE107 pKa = 4.13VEE109 pKa = 4.64DD110 pKa = 3.69SDD112 pKa = 4.32KK113 pKa = 10.63EE114 pKa = 4.14ARR116 pKa = 11.84EE117 pKa = 3.61IAYY120 pKa = 10.24GDD122 pKa = 4.16TIDD125 pKa = 5.02KK126 pKa = 11.06YY127 pKa = 9.96IDD129 pKa = 3.23KK130 pKa = 10.33RR131 pKa = 11.84AKK133 pKa = 8.97QLKK136 pKa = 8.43EE137 pKa = 3.37QATFDD142 pKa = 3.8EE143 pKa = 5.04LVDD146 pKa = 3.93KK147 pKa = 11.06VALYY151 pKa = 10.43EE152 pKa = 4.89SEE154 pKa = 4.11LLDD157 pKa = 3.57YY158 pKa = 11.28AEE160 pKa = 5.48RR161 pKa = 11.84LLSDD165 pKa = 4.64DD166 pKa = 4.48PLIADD171 pKa = 3.97SEE173 pKa = 4.7TAIGTLEE180 pKa = 3.99MLDD183 pKa = 3.96NEE185 pKa = 5.27AIDD188 pKa = 4.13LFKK191 pKa = 11.29SLDD194 pKa = 3.42IDD196 pKa = 3.65NEE198 pKa = 4.42YY199 pKa = 10.91QGLEE203 pKa = 4.14YY204 pKa = 11.01YY205 pKa = 8.17DD206 pKa = 3.56TSLNKK211 pKa = 9.96EE212 pKa = 4.17DD213 pKa = 3.66
Molecular weight: 24.72 kDa
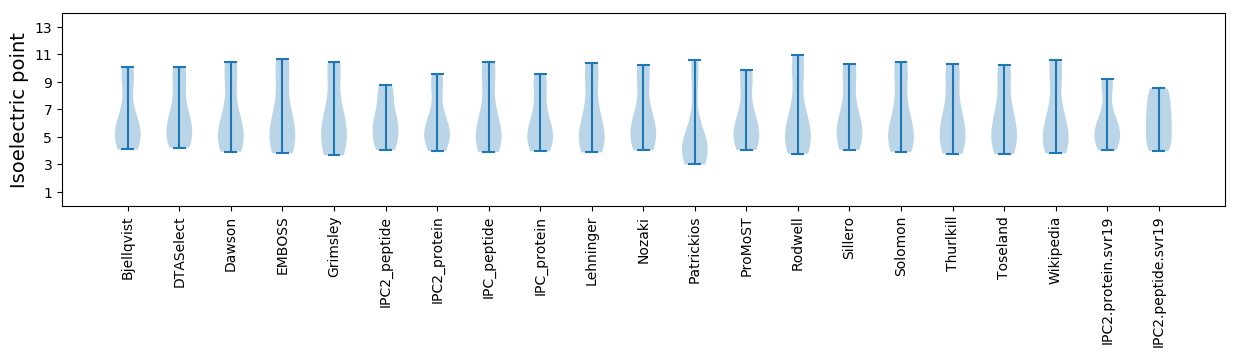
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D5ZNB4|A0A4D5ZNB4_9VIRU Uncharacterized protein OS=Streptococcus satellite phage Javan285 OX=2558607 GN=JavanS285_0009 PE=4 SV=1
MM1 pKa = 7.39FLHH4 pKa = 6.8FPIILDD10 pKa = 4.48GIIEE14 pKa = 4.21ADD16 pKa = 3.38EE17 pKa = 4.25TFFAISYY24 pKa = 9.35KK25 pKa = 10.68GNHH28 pKa = 6.13SKK30 pKa = 10.92SKK32 pKa = 8.95TFAMPRR38 pKa = 11.84KK39 pKa = 8.88AYY41 pKa = 9.67KK42 pKa = 9.82RR43 pKa = 11.84GHH45 pKa = 4.97STHH48 pKa = 6.36IRR50 pKa = 11.84GLSQEE55 pKa = 4.4KK56 pKa = 10.23VCVPCAVNRR65 pKa = 11.84NGLSISKK72 pKa = 8.26ITNTGRR78 pKa = 11.84VSTRR82 pKa = 11.84DD83 pKa = 3.09LHH85 pKa = 7.18HH86 pKa = 7.14IYY88 pKa = 10.55DD89 pKa = 4.01GRR91 pKa = 11.84IKK93 pKa = 10.08TNSTLVTDD101 pKa = 4.3KK102 pKa = 10.3MNSYY106 pKa = 10.45VRR108 pKa = 11.84FTNANGIDD116 pKa = 4.04LVQLKK121 pKa = 8.99TGKK124 pKa = 9.91AKK126 pKa = 10.54KK127 pKa = 10.1GIYY130 pKa = 9.6NIQHH134 pKa = 5.91INSYY138 pKa = 10.3HH139 pKa = 5.54SQLKK143 pKa = 9.97RR144 pKa = 11.84FMRR147 pKa = 11.84GFNGVSTKK155 pKa = 10.69YY156 pKa = 10.69LNNYY160 pKa = 7.83LVWNNLVNYY169 pKa = 9.92AKK171 pKa = 10.47EE172 pKa = 3.76SDD174 pKa = 3.53MEE176 pKa = 4.07KK177 pKa = 10.79RR178 pKa = 11.84NIFLTFVLATLKK190 pKa = 9.45TAKK193 pKa = 10.3CRR195 pKa = 11.84DD196 pKa = 3.51LSNRR200 pKa = 11.84PAVPLVAA207 pKa = 5.64
MM1 pKa = 7.39FLHH4 pKa = 6.8FPIILDD10 pKa = 4.48GIIEE14 pKa = 4.21ADD16 pKa = 3.38EE17 pKa = 4.25TFFAISYY24 pKa = 9.35KK25 pKa = 10.68GNHH28 pKa = 6.13SKK30 pKa = 10.92SKK32 pKa = 8.95TFAMPRR38 pKa = 11.84KK39 pKa = 8.88AYY41 pKa = 9.67KK42 pKa = 9.82RR43 pKa = 11.84GHH45 pKa = 4.97STHH48 pKa = 6.36IRR50 pKa = 11.84GLSQEE55 pKa = 4.4KK56 pKa = 10.23VCVPCAVNRR65 pKa = 11.84NGLSISKK72 pKa = 8.26ITNTGRR78 pKa = 11.84VSTRR82 pKa = 11.84DD83 pKa = 3.09LHH85 pKa = 7.18HH86 pKa = 7.14IYY88 pKa = 10.55DD89 pKa = 4.01GRR91 pKa = 11.84IKK93 pKa = 10.08TNSTLVTDD101 pKa = 4.3KK102 pKa = 10.3MNSYY106 pKa = 10.45VRR108 pKa = 11.84FTNANGIDD116 pKa = 4.04LVQLKK121 pKa = 8.99TGKK124 pKa = 9.91AKK126 pKa = 10.54KK127 pKa = 10.1GIYY130 pKa = 9.6NIQHH134 pKa = 5.91INSYY138 pKa = 10.3HH139 pKa = 5.54SQLKK143 pKa = 9.97RR144 pKa = 11.84FMRR147 pKa = 11.84GFNGVSTKK155 pKa = 10.69YY156 pKa = 10.69LNNYY160 pKa = 7.83LVWNNLVNYY169 pKa = 9.92AKK171 pKa = 10.47EE172 pKa = 3.76SDD174 pKa = 3.53MEE176 pKa = 4.07KK177 pKa = 10.79RR178 pKa = 11.84NIFLTFVLATLKK190 pKa = 9.45TAKK193 pKa = 10.3CRR195 pKa = 11.84DD196 pKa = 3.51LSNRR200 pKa = 11.84PAVPLVAA207 pKa = 5.64
Molecular weight: 23.56 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2731 |
52 |
468 |
170.7 |
19.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.163 ± 0.541 | 0.696 ± 0.211 |
6.225 ± 0.718 | 8.275 ± 0.875 |
3.881 ± 0.368 | 4.76 ± 0.322 |
2.417 ± 0.594 | 7.653 ± 0.549 |
8.825 ± 0.545 | 9.667 ± 0.711 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.417 ± 0.254 | 5.785 ± 0.524 |
2.673 ± 0.344 | 3.881 ± 0.416 |
5.126 ± 0.611 | 4.65 ± 0.446 |
6.518 ± 0.47 | 5.419 ± 0.428 |
0.952 ± 0.16 | 5.016 ± 0.456 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
