
Rhizobium smilacinae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Rhizobiaceae; Rhizobium/Agrobacterium group; Rhizobium
Average proteome isoelectric point is 6.39
Get precalculated fractions of proteins
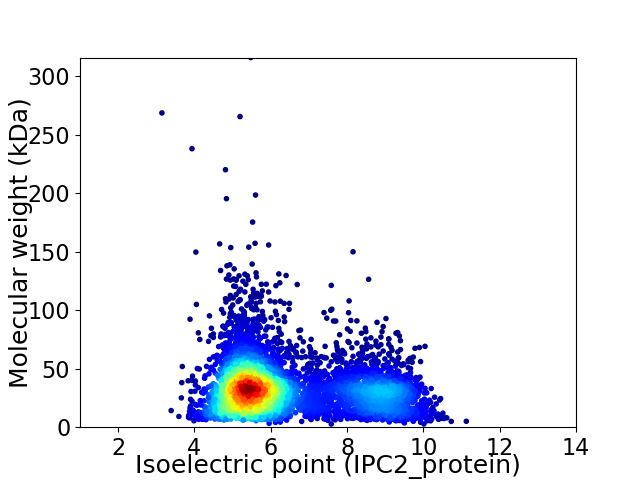
Virtual 2D-PAGE plot for 5526 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5C4XJ63|A0A5C4XJ63_9RHIZ Inositol-1-monophosphatase OS=Rhizobium smilacinae OX=1395944 GN=FHP24_15605 PE=3 SV=1
MM1 pKa = 7.35TNSISNALGVALYY14 pKa = 10.71YY15 pKa = 10.76SGSSKK20 pKa = 10.9AWFSATGSGPIQYY33 pKa = 7.86GTAGNDD39 pKa = 3.8SMWGDD44 pKa = 3.38SSVNVTMTGGTGDD57 pKa = 4.7DD58 pKa = 2.99IYY60 pKa = 11.77YY61 pKa = 10.53LYY63 pKa = 10.76SDD65 pKa = 3.96KK66 pKa = 11.22NRR68 pKa = 11.84AEE70 pKa = 4.16EE71 pKa = 4.07KK72 pKa = 10.52AGEE75 pKa = 4.3GVDD78 pKa = 5.46TIDD81 pKa = 2.52TWMSYY86 pKa = 7.26TLPEE90 pKa = 3.96NFEE93 pKa = 4.28NLRR96 pKa = 11.84VTGDD100 pKa = 2.87NRR102 pKa = 11.84YY103 pKa = 10.34AFGNSADD110 pKa = 4.02NIITGSSSRR119 pKa = 11.84QTIDD123 pKa = 2.75GGAGNDD129 pKa = 3.49VLIGGGGADD138 pKa = 3.41TFIISKK144 pKa = 11.04GNGTDD149 pKa = 4.64LIADD153 pKa = 4.73FSSDD157 pKa = 3.78DD158 pKa = 3.98RR159 pKa = 11.84IRR161 pKa = 11.84LDD163 pKa = 3.67GYY165 pKa = 11.04GVTSFDD171 pKa = 3.53QLISQSSQQGNDD183 pKa = 2.59LWLNLGNGEE192 pKa = 4.46SVVLAGTTADD202 pKa = 5.27DD203 pKa = 4.19LSADD207 pKa = 3.67QFEE210 pKa = 5.15LNLDD214 pKa = 3.77RR215 pKa = 11.84SSLTLSFNEE224 pKa = 4.62DD225 pKa = 3.54FNSLSLFDD233 pKa = 4.41GQDD236 pKa = 3.8GVWEE240 pKa = 4.19AKK242 pKa = 10.07YY243 pKa = 9.14WWAPEE248 pKa = 4.0KK249 pKa = 10.69GASLHH254 pKa = 6.36TNGEE258 pKa = 4.26QQWYY262 pKa = 9.67VNPAYY267 pKa = 10.53DD268 pKa = 4.03PTASANPFSVTDD280 pKa = 3.93GVLTIRR286 pKa = 11.84AEE288 pKa = 4.03QTPDD292 pKa = 3.24ALSGEE297 pKa = 4.53VEE299 pKa = 4.55GYY301 pKa = 10.79DD302 pKa = 3.66YY303 pKa = 11.18TSGMLTTHH311 pKa = 7.26ASFAQTYY318 pKa = 9.42GYY320 pKa = 10.77FEE322 pKa = 4.72IRR324 pKa = 11.84ADD326 pKa = 3.68MPDD329 pKa = 3.47EE330 pKa = 4.14QGAWPAFWLLPEE342 pKa = 5.2DD343 pKa = 4.82GSWPPEE349 pKa = 3.64LDD351 pKa = 3.61VIEE354 pKa = 5.03MRR356 pKa = 11.84GQDD359 pKa = 4.05PNTLIMSAHH368 pKa = 6.06SNEE371 pKa = 4.43TGQQTSVIKK380 pKa = 10.72NVAVTDD386 pKa = 3.61TEE388 pKa = 4.62GFHH391 pKa = 6.81TYY393 pKa = 10.64GLLWDD398 pKa = 4.42EE399 pKa = 5.7DD400 pKa = 4.33HH401 pKa = 6.44ITWYY405 pKa = 10.65FDD407 pKa = 3.53DD408 pKa = 4.01VAVAQTDD415 pKa = 4.06TPADD419 pKa = 3.63MHH421 pKa = 7.35DD422 pKa = 3.79PMYY425 pKa = 10.22MIVNLAVGGMAGTPSDD441 pKa = 4.16EE442 pKa = 4.71LPDD445 pKa = 4.05GSEE448 pKa = 3.86MNIDD452 pKa = 3.97YY453 pKa = 10.66IRR455 pKa = 11.84AYY457 pKa = 10.96SLDD460 pKa = 3.9DD461 pKa = 4.07LEE463 pKa = 5.54ASNTASAVQPSEE475 pKa = 3.74WLLL478 pKa = 3.6
MM1 pKa = 7.35TNSISNALGVALYY14 pKa = 10.71YY15 pKa = 10.76SGSSKK20 pKa = 10.9AWFSATGSGPIQYY33 pKa = 7.86GTAGNDD39 pKa = 3.8SMWGDD44 pKa = 3.38SSVNVTMTGGTGDD57 pKa = 4.7DD58 pKa = 2.99IYY60 pKa = 11.77YY61 pKa = 10.53LYY63 pKa = 10.76SDD65 pKa = 3.96KK66 pKa = 11.22NRR68 pKa = 11.84AEE70 pKa = 4.16EE71 pKa = 4.07KK72 pKa = 10.52AGEE75 pKa = 4.3GVDD78 pKa = 5.46TIDD81 pKa = 2.52TWMSYY86 pKa = 7.26TLPEE90 pKa = 3.96NFEE93 pKa = 4.28NLRR96 pKa = 11.84VTGDD100 pKa = 2.87NRR102 pKa = 11.84YY103 pKa = 10.34AFGNSADD110 pKa = 4.02NIITGSSSRR119 pKa = 11.84QTIDD123 pKa = 2.75GGAGNDD129 pKa = 3.49VLIGGGGADD138 pKa = 3.41TFIISKK144 pKa = 11.04GNGTDD149 pKa = 4.64LIADD153 pKa = 4.73FSSDD157 pKa = 3.78DD158 pKa = 3.98RR159 pKa = 11.84IRR161 pKa = 11.84LDD163 pKa = 3.67GYY165 pKa = 11.04GVTSFDD171 pKa = 3.53QLISQSSQQGNDD183 pKa = 2.59LWLNLGNGEE192 pKa = 4.46SVVLAGTTADD202 pKa = 5.27DD203 pKa = 4.19LSADD207 pKa = 3.67QFEE210 pKa = 5.15LNLDD214 pKa = 3.77RR215 pKa = 11.84SSLTLSFNEE224 pKa = 4.62DD225 pKa = 3.54FNSLSLFDD233 pKa = 4.41GQDD236 pKa = 3.8GVWEE240 pKa = 4.19AKK242 pKa = 10.07YY243 pKa = 9.14WWAPEE248 pKa = 4.0KK249 pKa = 10.69GASLHH254 pKa = 6.36TNGEE258 pKa = 4.26QQWYY262 pKa = 9.67VNPAYY267 pKa = 10.53DD268 pKa = 4.03PTASANPFSVTDD280 pKa = 3.93GVLTIRR286 pKa = 11.84AEE288 pKa = 4.03QTPDD292 pKa = 3.24ALSGEE297 pKa = 4.53VEE299 pKa = 4.55GYY301 pKa = 10.79DD302 pKa = 3.66YY303 pKa = 11.18TSGMLTTHH311 pKa = 7.26ASFAQTYY318 pKa = 9.42GYY320 pKa = 10.77FEE322 pKa = 4.72IRR324 pKa = 11.84ADD326 pKa = 3.68MPDD329 pKa = 3.47EE330 pKa = 4.14QGAWPAFWLLPEE342 pKa = 5.2DD343 pKa = 4.82GSWPPEE349 pKa = 3.64LDD351 pKa = 3.61VIEE354 pKa = 5.03MRR356 pKa = 11.84GQDD359 pKa = 4.05PNTLIMSAHH368 pKa = 6.06SNEE371 pKa = 4.43TGQQTSVIKK380 pKa = 10.72NVAVTDD386 pKa = 3.61TEE388 pKa = 4.62GFHH391 pKa = 6.81TYY393 pKa = 10.64GLLWDD398 pKa = 4.42EE399 pKa = 5.7DD400 pKa = 4.33HH401 pKa = 6.44ITWYY405 pKa = 10.65FDD407 pKa = 3.53DD408 pKa = 4.01VAVAQTDD415 pKa = 4.06TPADD419 pKa = 3.63MHH421 pKa = 7.35DD422 pKa = 3.79PMYY425 pKa = 10.22MIVNLAVGGMAGTPSDD441 pKa = 4.16EE442 pKa = 4.71LPDD445 pKa = 4.05GSEE448 pKa = 3.86MNIDD452 pKa = 3.97YY453 pKa = 10.66IRR455 pKa = 11.84AYY457 pKa = 10.96SLDD460 pKa = 3.9DD461 pKa = 4.07LEE463 pKa = 5.54ASNTASAVQPSEE475 pKa = 3.74WLLL478 pKa = 3.6
Molecular weight: 51.83 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5C4XSC8|A0A5C4XSC8_9RHIZ Phosphonate ABC transporter substrate-binding protein OS=Rhizobium smilacinae OX=1395944 GN=FHP24_08570 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.22GGRR28 pKa = 11.84KK29 pKa = 9.06VLSARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84ASLSAA44 pKa = 3.92
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.22GGRR28 pKa = 11.84KK29 pKa = 9.06VLSARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84ASLSAA44 pKa = 3.92
Molecular weight: 5.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1739106 |
24 |
2820 |
314.7 |
34.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.527 ± 0.04 | 0.765 ± 0.011 |
5.724 ± 0.027 | 5.723 ± 0.031 |
3.964 ± 0.026 | 8.302 ± 0.03 |
1.971 ± 0.017 | 5.927 ± 0.024 |
3.826 ± 0.028 | 9.945 ± 0.033 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.616 ± 0.015 | 2.899 ± 0.015 |
4.777 ± 0.026 | 3.173 ± 0.02 |
6.422 ± 0.029 | 6.034 ± 0.027 |
5.472 ± 0.03 | 7.334 ± 0.026 |
1.257 ± 0.013 | 2.342 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
