
McMurdo Ice Shelf pond-associated circular DNA virus-7
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 8.48
Get precalculated fractions of proteins
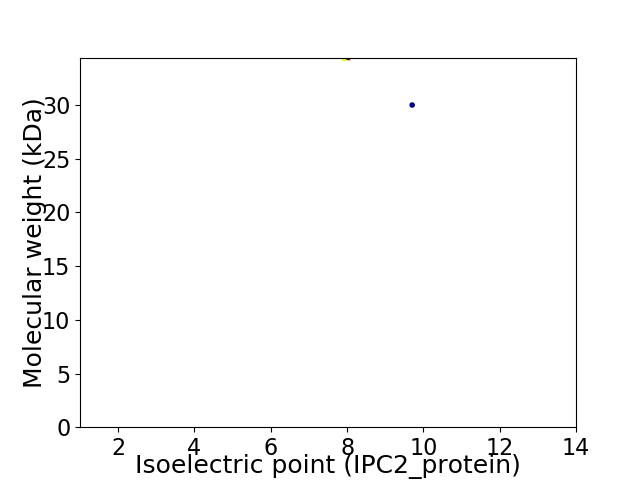
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A075M3Q4|A0A075M3Q4_9VIRU Putative capsid protein OS=McMurdo Ice Shelf pond-associated circular DNA virus-7 OX=1521391 PE=4 SV=1
MM1 pKa = 7.26PQPRR5 pKa = 11.84MKK7 pKa = 10.82DD8 pKa = 3.34NEE10 pKa = 4.21VKK12 pKa = 10.55KK13 pKa = 10.98VFNALVFTDD22 pKa = 4.28FSLYY26 pKa = 10.49KK27 pKa = 8.75WTLEE31 pKa = 3.96DD32 pKa = 4.04LATHH36 pKa = 7.3KK37 pKa = 10.29IQYY40 pKa = 8.85LAYY43 pKa = 9.81GLEE46 pKa = 4.35TCPEE50 pKa = 4.25TKK52 pKa = 9.94KK53 pKa = 9.98QHH55 pKa = 5.39HH56 pKa = 6.07QGWLYY61 pKa = 11.26ASTDD65 pKa = 3.31RR66 pKa = 11.84KK67 pKa = 10.05RR68 pKa = 11.84SFMAWKK74 pKa = 10.03KK75 pKa = 9.95HH76 pKa = 4.1FQLLGLDD83 pKa = 3.39MHH85 pKa = 6.87YY86 pKa = 11.03EE87 pKa = 3.96AMQGNFSQNTKK98 pKa = 10.24YY99 pKa = 10.21ISKK102 pKa = 10.1EE103 pKa = 3.93GQLTEE108 pKa = 5.34LGVKK112 pKa = 10.17PMGSGKK118 pKa = 9.99KK119 pKa = 8.32RR120 pKa = 11.84TLLEE124 pKa = 3.8YY125 pKa = 10.37KK126 pKa = 10.33KK127 pKa = 10.38QIEE130 pKa = 4.39AGAHH134 pKa = 4.94VLDD137 pKa = 4.65IAEE140 pKa = 4.38EE141 pKa = 4.21DD142 pKa = 3.7EE143 pKa = 4.54KK144 pKa = 11.35FGTFLQYY151 pKa = 10.72RR152 pKa = 11.84SGLNAYY158 pKa = 9.89ADD160 pKa = 3.53HH161 pKa = 6.52QRR163 pKa = 11.84FKK165 pKa = 10.99RR166 pKa = 11.84IRR168 pKa = 11.84TSRR171 pKa = 11.84EE172 pKa = 3.35KK173 pKa = 9.83PEE175 pKa = 3.64VYY177 pKa = 10.0IRR179 pKa = 11.84VGPPGTGKK187 pKa = 10.36SRR189 pKa = 11.84WLDD192 pKa = 3.22EE193 pKa = 4.32TYY195 pKa = 11.2GFDD198 pKa = 3.59KK199 pKa = 10.57WIEE202 pKa = 4.06APDD205 pKa = 3.78NSGKK209 pKa = 9.76WFDD212 pKa = 3.98GCDD215 pKa = 3.25LADD218 pKa = 3.86IIVFNDD224 pKa = 3.6VGPGSCPPLDD234 pKa = 3.44IFKK237 pKa = 10.23KK238 pKa = 10.6LCDD241 pKa = 3.62RR242 pKa = 11.84YY243 pKa = 9.89PFRR246 pKa = 11.84GAVKK250 pKa = 10.51GGFIWLKK257 pKa = 9.67PKK259 pKa = 10.48IIVFTSNSYY268 pKa = 5.38WTEE271 pKa = 3.29WFGPDD276 pKa = 4.36LAPLDD281 pKa = 3.73RR282 pKa = 11.84EE283 pKa = 4.34AIEE286 pKa = 4.27RR287 pKa = 11.84RR288 pKa = 11.84ITSTICVEE296 pKa = 4.09
MM1 pKa = 7.26PQPRR5 pKa = 11.84MKK7 pKa = 10.82DD8 pKa = 3.34NEE10 pKa = 4.21VKK12 pKa = 10.55KK13 pKa = 10.98VFNALVFTDD22 pKa = 4.28FSLYY26 pKa = 10.49KK27 pKa = 8.75WTLEE31 pKa = 3.96DD32 pKa = 4.04LATHH36 pKa = 7.3KK37 pKa = 10.29IQYY40 pKa = 8.85LAYY43 pKa = 9.81GLEE46 pKa = 4.35TCPEE50 pKa = 4.25TKK52 pKa = 9.94KK53 pKa = 9.98QHH55 pKa = 5.39HH56 pKa = 6.07QGWLYY61 pKa = 11.26ASTDD65 pKa = 3.31RR66 pKa = 11.84KK67 pKa = 10.05RR68 pKa = 11.84SFMAWKK74 pKa = 10.03KK75 pKa = 9.95HH76 pKa = 4.1FQLLGLDD83 pKa = 3.39MHH85 pKa = 6.87YY86 pKa = 11.03EE87 pKa = 3.96AMQGNFSQNTKK98 pKa = 10.24YY99 pKa = 10.21ISKK102 pKa = 10.1EE103 pKa = 3.93GQLTEE108 pKa = 5.34LGVKK112 pKa = 10.17PMGSGKK118 pKa = 9.99KK119 pKa = 8.32RR120 pKa = 11.84TLLEE124 pKa = 3.8YY125 pKa = 10.37KK126 pKa = 10.33KK127 pKa = 10.38QIEE130 pKa = 4.39AGAHH134 pKa = 4.94VLDD137 pKa = 4.65IAEE140 pKa = 4.38EE141 pKa = 4.21DD142 pKa = 3.7EE143 pKa = 4.54KK144 pKa = 11.35FGTFLQYY151 pKa = 10.72RR152 pKa = 11.84SGLNAYY158 pKa = 9.89ADD160 pKa = 3.53HH161 pKa = 6.52QRR163 pKa = 11.84FKK165 pKa = 10.99RR166 pKa = 11.84IRR168 pKa = 11.84TSRR171 pKa = 11.84EE172 pKa = 3.35KK173 pKa = 9.83PEE175 pKa = 3.64VYY177 pKa = 10.0IRR179 pKa = 11.84VGPPGTGKK187 pKa = 10.36SRR189 pKa = 11.84WLDD192 pKa = 3.22EE193 pKa = 4.32TYY195 pKa = 11.2GFDD198 pKa = 3.59KK199 pKa = 10.57WIEE202 pKa = 4.06APDD205 pKa = 3.78NSGKK209 pKa = 9.76WFDD212 pKa = 3.98GCDD215 pKa = 3.25LADD218 pKa = 3.86IIVFNDD224 pKa = 3.6VGPGSCPPLDD234 pKa = 3.44IFKK237 pKa = 10.23KK238 pKa = 10.6LCDD241 pKa = 3.62RR242 pKa = 11.84YY243 pKa = 9.89PFRR246 pKa = 11.84GAVKK250 pKa = 10.51GGFIWLKK257 pKa = 9.67PKK259 pKa = 10.48IIVFTSNSYY268 pKa = 5.38WTEE271 pKa = 3.29WFGPDD276 pKa = 4.36LAPLDD281 pKa = 3.73RR282 pKa = 11.84EE283 pKa = 4.34AIEE286 pKa = 4.27RR287 pKa = 11.84RR288 pKa = 11.84ITSTICVEE296 pKa = 4.09
Molecular weight: 34.32 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A075M3Q4|A0A075M3Q4_9VIRU Putative capsid protein OS=McMurdo Ice Shelf pond-associated circular DNA virus-7 OX=1521391 PE=4 SV=1
MM1 pKa = 6.64VFKK4 pKa = 10.77RR5 pKa = 11.84KK6 pKa = 9.44FSSKK10 pKa = 9.9RR11 pKa = 11.84MGRR14 pKa = 11.84GSIRR18 pKa = 11.84KK19 pKa = 8.59PFIKK23 pKa = 10.25RR24 pKa = 11.84GTRR27 pKa = 11.84RR28 pKa = 11.84VGSAIRR34 pKa = 11.84NLAKK38 pKa = 10.08KK39 pKa = 9.61VRR41 pKa = 11.84RR42 pKa = 11.84ITSTIEE48 pKa = 3.8TKK50 pKa = 10.74SGVEE54 pKa = 4.1QITDD58 pKa = 3.87NINLRR63 pKa = 11.84HH64 pKa = 5.82NNQVTVSSAVLEE76 pKa = 4.6TQTGTLDD83 pKa = 3.57NEE85 pKa = 4.49NSRR88 pKa = 11.84GMRR91 pKa = 11.84IGDD94 pKa = 4.56KK95 pKa = 9.4ITLSNVQFTMMFEE108 pKa = 4.28LNEE111 pKa = 4.42AYY113 pKa = 9.69TDD115 pKa = 3.36VTLRR119 pKa = 11.84LMVVRR124 pKa = 11.84AAKK127 pKa = 10.4NDD129 pKa = 3.29IPNITDD135 pKa = 3.41MFHH138 pKa = 6.4GASGNKK144 pKa = 9.01MLDD147 pKa = 3.11TFNNEE152 pKa = 4.07RR153 pKa = 11.84YY154 pKa = 10.48SMLYY158 pKa = 8.88QKK160 pKa = 9.91YY161 pKa = 9.09VRR163 pKa = 11.84LRR165 pKa = 11.84SNPTAAVPSGTQFPGSGTYY184 pKa = 9.61QGPNQIMSRR193 pKa = 11.84ATKK196 pKa = 9.12IVKK199 pKa = 10.22FSVPGRR205 pKa = 11.84KK206 pKa = 8.91FSRR209 pKa = 11.84SGIIQYY215 pKa = 10.06QNNDD219 pKa = 3.27VQPKK223 pKa = 9.26FFDD226 pKa = 3.82YY227 pKa = 11.18YY228 pKa = 10.48FIVYY232 pKa = 9.86AYY234 pKa = 11.17SNFATVDD241 pKa = 3.53SGPVVYY247 pKa = 10.5NVAHH251 pKa = 6.75INDD254 pKa = 4.82CIIRR258 pKa = 11.84MHH260 pKa = 6.43YY261 pKa = 10.39KK262 pKa = 10.47DD263 pKa = 3.44AA264 pKa = 5.6
MM1 pKa = 6.64VFKK4 pKa = 10.77RR5 pKa = 11.84KK6 pKa = 9.44FSSKK10 pKa = 9.9RR11 pKa = 11.84MGRR14 pKa = 11.84GSIRR18 pKa = 11.84KK19 pKa = 8.59PFIKK23 pKa = 10.25RR24 pKa = 11.84GTRR27 pKa = 11.84RR28 pKa = 11.84VGSAIRR34 pKa = 11.84NLAKK38 pKa = 10.08KK39 pKa = 9.61VRR41 pKa = 11.84RR42 pKa = 11.84ITSTIEE48 pKa = 3.8TKK50 pKa = 10.74SGVEE54 pKa = 4.1QITDD58 pKa = 3.87NINLRR63 pKa = 11.84HH64 pKa = 5.82NNQVTVSSAVLEE76 pKa = 4.6TQTGTLDD83 pKa = 3.57NEE85 pKa = 4.49NSRR88 pKa = 11.84GMRR91 pKa = 11.84IGDD94 pKa = 4.56KK95 pKa = 9.4ITLSNVQFTMMFEE108 pKa = 4.28LNEE111 pKa = 4.42AYY113 pKa = 9.69TDD115 pKa = 3.36VTLRR119 pKa = 11.84LMVVRR124 pKa = 11.84AAKK127 pKa = 10.4NDD129 pKa = 3.29IPNITDD135 pKa = 3.41MFHH138 pKa = 6.4GASGNKK144 pKa = 9.01MLDD147 pKa = 3.11TFNNEE152 pKa = 4.07RR153 pKa = 11.84YY154 pKa = 10.48SMLYY158 pKa = 8.88QKK160 pKa = 9.91YY161 pKa = 9.09VRR163 pKa = 11.84LRR165 pKa = 11.84SNPTAAVPSGTQFPGSGTYY184 pKa = 9.61QGPNQIMSRR193 pKa = 11.84ATKK196 pKa = 9.12IVKK199 pKa = 10.22FSVPGRR205 pKa = 11.84KK206 pKa = 8.91FSRR209 pKa = 11.84SGIIQYY215 pKa = 10.06QNNDD219 pKa = 3.27VQPKK223 pKa = 9.26FFDD226 pKa = 3.82YY227 pKa = 11.18YY228 pKa = 10.48FIVYY232 pKa = 9.86AYY234 pKa = 11.17SNFATVDD241 pKa = 3.53SGPVVYY247 pKa = 10.5NVAHH251 pKa = 6.75INDD254 pKa = 4.82CIIRR258 pKa = 11.84MHH260 pKa = 6.43YY261 pKa = 10.39KK262 pKa = 10.47DD263 pKa = 3.44AA264 pKa = 5.6
Molecular weight: 30.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
560 |
264 |
296 |
280.0 |
32.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.357 ± 0.035 | 1.071 ± 0.445 |
5.536 ± 0.636 | 4.821 ± 1.395 |
5.536 ± 0.15 | 7.143 ± 0.452 |
1.964 ± 0.289 | 6.25 ± 0.609 |
8.036 ± 1.026 | 6.25 ± 1.339 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.036 ± 0.727 | 5.179 ± 1.784 |
4.286 ± 0.563 | 3.929 ± 0.153 |
6.786 ± 0.995 | 6.25 ± 1.096 |
6.607 ± 0.623 | 5.893 ± 1.325 |
1.607 ± 1.033 | 4.464 ± 0.052 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
