
Nicotiana attenuata (Coyote tobacco)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Nicotianoideae; Nicotianeae; Nicotiana
Average proteome isoelectric point is 6.74
Get precalculated fractions of proteins
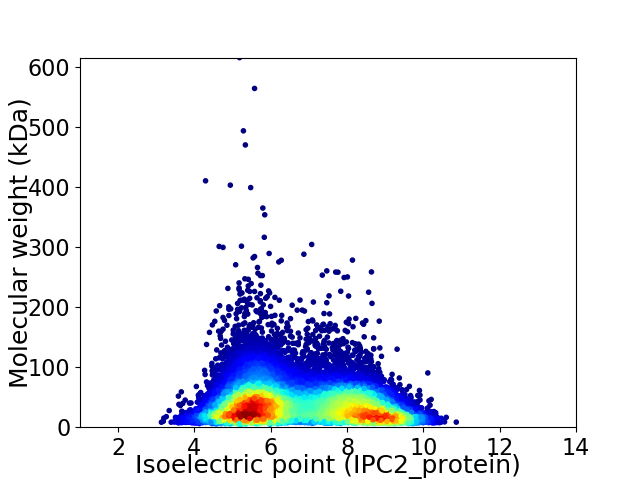
Virtual 2D-PAGE plot for 32840 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1J6L408|A0A1J6L408_NICAT Protein mut11 OS=Nicotiana attenuata OX=49451 GN=MUT11_0 PE=3 SV=1
MM1 pKa = 7.14ATLVPEE7 pKa = 4.25MEE9 pKa = 3.88NWACINGCNDD19 pKa = 4.24LGNFDD24 pKa = 4.72QSQIDD29 pKa = 3.9CALLMSLLDD38 pKa = 4.17DD39 pKa = 4.02SQLGVEE45 pKa = 4.93DD46 pKa = 3.43EE47 pKa = 4.71HH48 pKa = 8.58DD49 pKa = 3.88NKK51 pKa = 10.81RR52 pKa = 11.84LTSVIRR58 pKa = 11.84SLEE61 pKa = 3.91AEE63 pKa = 4.03IDD65 pKa = 3.74DD66 pKa = 3.63QQVINGHH73 pKa = 6.81DD74 pKa = 3.73SFQDD78 pKa = 3.41SQGRR82 pKa = 11.84NSGEE86 pKa = 3.85DD87 pKa = 3.05WQLWEE92 pKa = 4.4DD93 pKa = 3.71SQYY96 pKa = 11.49KK97 pKa = 9.91SQDD100 pKa = 3.31FLEE103 pKa = 4.86SEE105 pKa = 4.28GLDD108 pKa = 4.12LNWMALEE115 pKa = 4.09MEE117 pKa = 4.82IAPSSTSDD125 pKa = 5.77DD126 pKa = 3.28MNFWYY131 pKa = 10.21NHH133 pKa = 5.29QVHH136 pKa = 7.04DD137 pKa = 3.8MCKK140 pKa = 8.41YY141 pKa = 9.62GYY143 pKa = 10.53AAFSLEE149 pKa = 4.3EE150 pKa = 4.09NDD152 pKa = 3.74HH153 pKa = 6.19NSWGQEE159 pKa = 3.96QNSSALTMQGG169 pKa = 2.9
MM1 pKa = 7.14ATLVPEE7 pKa = 4.25MEE9 pKa = 3.88NWACINGCNDD19 pKa = 4.24LGNFDD24 pKa = 4.72QSQIDD29 pKa = 3.9CALLMSLLDD38 pKa = 4.17DD39 pKa = 4.02SQLGVEE45 pKa = 4.93DD46 pKa = 3.43EE47 pKa = 4.71HH48 pKa = 8.58DD49 pKa = 3.88NKK51 pKa = 10.81RR52 pKa = 11.84LTSVIRR58 pKa = 11.84SLEE61 pKa = 3.91AEE63 pKa = 4.03IDD65 pKa = 3.74DD66 pKa = 3.63QQVINGHH73 pKa = 6.81DD74 pKa = 3.73SFQDD78 pKa = 3.41SQGRR82 pKa = 11.84NSGEE86 pKa = 3.85DD87 pKa = 3.05WQLWEE92 pKa = 4.4DD93 pKa = 3.71SQYY96 pKa = 11.49KK97 pKa = 9.91SQDD100 pKa = 3.31FLEE103 pKa = 4.86SEE105 pKa = 4.28GLDD108 pKa = 4.12LNWMALEE115 pKa = 4.09MEE117 pKa = 4.82IAPSSTSDD125 pKa = 5.77DD126 pKa = 3.28MNFWYY131 pKa = 10.21NHH133 pKa = 5.29QVHH136 pKa = 7.04DD137 pKa = 3.8MCKK140 pKa = 8.41YY141 pKa = 9.62GYY143 pKa = 10.53AAFSLEE149 pKa = 4.3EE150 pKa = 4.09NDD152 pKa = 3.74HH153 pKa = 6.19NSWGQEE159 pKa = 3.96QNSSALTMQGG169 pKa = 2.9
Molecular weight: 19.29 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A314L286|A0A314L286_NICAT Uncharacterized protein OS=Nicotiana attenuata OX=49451 GN=A4A49_04192 PE=4 SV=1
MM1 pKa = 7.71KK2 pKa = 10.58GRR4 pKa = 11.84GFRR7 pKa = 11.84LGRR10 pKa = 11.84RR11 pKa = 11.84LVRR14 pKa = 11.84VFSCFIRR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84TNGRR27 pKa = 11.84VGDD30 pKa = 3.7KK31 pKa = 10.32RR32 pKa = 11.84LRR34 pKa = 11.84SQGCGSRR41 pKa = 11.84AISKK45 pKa = 10.14LSKK48 pKa = 10.18CLRR51 pKa = 11.84LLKK54 pKa = 10.52HH55 pKa = 5.54GAKK58 pKa = 10.1GICFAKK64 pKa = 10.3PNSGYY69 pKa = 10.19FRR71 pKa = 11.84VGQEE75 pKa = 4.26PIDD78 pKa = 4.14PKK80 pKa = 10.74QVSVPKK86 pKa = 10.36GHH88 pKa = 6.7LAVYY92 pKa = 9.99VGEE95 pKa = 5.41KK96 pKa = 9.61IDD98 pKa = 3.6DD99 pKa = 3.83TCRR102 pKa = 11.84VVVPVIYY109 pKa = 10.43FNHH112 pKa = 7.06PLFADD117 pKa = 4.33LLRR120 pKa = 11.84EE121 pKa = 4.01AEE123 pKa = 3.89MTYY126 pKa = 10.79GYY128 pKa = 9.8NHH130 pKa = 6.8SGGIQIPCRR139 pKa = 11.84ISEE142 pKa = 4.2FEE144 pKa = 4.09NVKK147 pKa = 10.59SRR149 pKa = 11.84IAATGGGGNCRR160 pKa = 11.84RR161 pKa = 4.05
MM1 pKa = 7.71KK2 pKa = 10.58GRR4 pKa = 11.84GFRR7 pKa = 11.84LGRR10 pKa = 11.84RR11 pKa = 11.84LVRR14 pKa = 11.84VFSCFIRR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84TNGRR27 pKa = 11.84VGDD30 pKa = 3.7KK31 pKa = 10.32RR32 pKa = 11.84LRR34 pKa = 11.84SQGCGSRR41 pKa = 11.84AISKK45 pKa = 10.14LSKK48 pKa = 10.18CLRR51 pKa = 11.84LLKK54 pKa = 10.52HH55 pKa = 5.54GAKK58 pKa = 10.1GICFAKK64 pKa = 10.3PNSGYY69 pKa = 10.19FRR71 pKa = 11.84VGQEE75 pKa = 4.26PIDD78 pKa = 4.14PKK80 pKa = 10.74QVSVPKK86 pKa = 10.36GHH88 pKa = 6.7LAVYY92 pKa = 9.99VGEE95 pKa = 5.41KK96 pKa = 9.61IDD98 pKa = 3.6DD99 pKa = 3.83TCRR102 pKa = 11.84VVVPVIYY109 pKa = 10.43FNHH112 pKa = 7.06PLFADD117 pKa = 4.33LLRR120 pKa = 11.84EE121 pKa = 4.01AEE123 pKa = 3.89MTYY126 pKa = 10.79GYY128 pKa = 9.8NHH130 pKa = 6.8SGGIQIPCRR139 pKa = 11.84ISEE142 pKa = 4.2FEE144 pKa = 4.09NVKK147 pKa = 10.59SRR149 pKa = 11.84IAATGGGGNCRR160 pKa = 11.84RR161 pKa = 4.05
Molecular weight: 17.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
12261239 |
45 |
5431 |
373.4 |
41.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.627 ± 0.013 | 1.843 ± 0.007 |
5.301 ± 0.01 | 6.561 ± 0.016 |
4.203 ± 0.009 | 6.324 ± 0.013 |
2.342 ± 0.007 | 5.507 ± 0.011 |
6.424 ± 0.013 | 9.477 ± 0.017 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.434 ± 0.006 | 4.749 ± 0.01 |
4.84 ± 0.014 | 3.806 ± 0.01 |
5.093 ± 0.011 | 8.864 ± 0.017 |
4.994 ± 0.008 | 6.489 ± 0.011 |
1.253 ± 0.005 | 2.869 ± 0.008 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
