
Circovirus-like genome DCCV-12
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 8.14
Get precalculated fractions of proteins
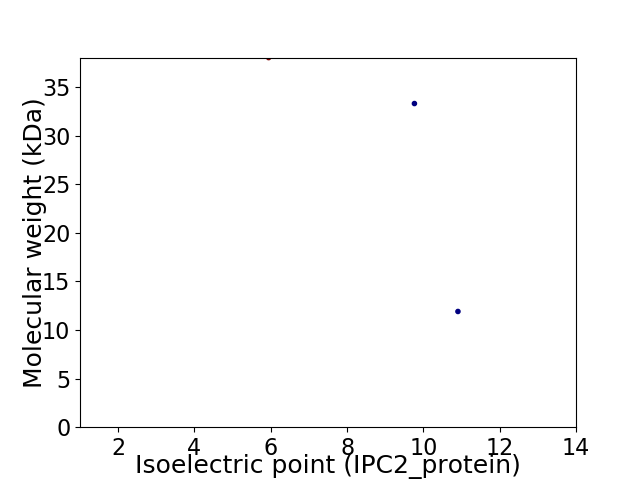
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A190WHF0|A0A190WHF0_9CIRC Uncharacterized protein OS=Circovirus-like genome DCCV-12 OX=1788440 PE=4 SV=1
MM1 pKa = 7.53PPKK4 pKa = 10.4AQFNRR9 pKa = 11.84FCQITKK15 pKa = 10.26KK16 pKa = 10.52RR17 pKa = 11.84GFCWTYY23 pKa = 10.59HH24 pKa = 7.02DD25 pKa = 4.68YY26 pKa = 11.56DD27 pKa = 4.73DD28 pKa = 6.15EE29 pKa = 4.82IVAHH33 pKa = 6.61IKK35 pKa = 10.92GLFDD39 pKa = 5.37RR40 pKa = 11.84PNGWVKK46 pKa = 10.57RR47 pKa = 11.84LVFNYY52 pKa = 7.44EE53 pKa = 3.54ICPTTGRR60 pKa = 11.84RR61 pKa = 11.84HH62 pKa = 5.42LQGCLYY68 pKa = 10.25MRR70 pKa = 11.84DD71 pKa = 3.49NQTWPKK77 pKa = 10.14LKK79 pKa = 10.61ADD81 pKa = 3.41IGLEE85 pKa = 4.18KK86 pKa = 10.33IHH88 pKa = 7.17LEE90 pKa = 3.72LAQNVEE96 pKa = 4.71ALDD99 pKa = 4.26KK100 pKa = 10.78YY101 pKa = 9.03CQKK104 pKa = 10.87SEE106 pKa = 4.19TRR108 pKa = 11.84DD109 pKa = 3.36PAHH112 pKa = 7.13PEE114 pKa = 3.99GVVLGVAPLTDD125 pKa = 3.34EE126 pKa = 5.21AKK128 pKa = 9.22GQKK131 pKa = 10.11GEE133 pKa = 4.06EE134 pKa = 4.3YY135 pKa = 10.63YY136 pKa = 10.81EE137 pKa = 4.38RR138 pKa = 11.84NILACMNGQQMDD150 pKa = 3.56ASAEE154 pKa = 3.97FNLRR158 pKa = 11.84NFEE161 pKa = 4.15YY162 pKa = 10.54AAEE165 pKa = 3.9ARR167 pKa = 11.84KK168 pKa = 9.6RR169 pKa = 11.84KK170 pKa = 9.86RR171 pKa = 11.84EE172 pKa = 3.92LASLDD177 pKa = 3.5EE178 pKa = 5.91LSFEE182 pKa = 4.01WHH184 pKa = 6.01YY185 pKa = 11.75GEE187 pKa = 4.75PFSGKK192 pKa = 6.61THH194 pKa = 5.96YY195 pKa = 10.01CRR197 pKa = 11.84KK198 pKa = 9.73IPGAFKK204 pKa = 10.13WNSKK208 pKa = 10.48AGWNNYY214 pKa = 8.25NDD216 pKa = 4.18EE217 pKa = 4.43EE218 pKa = 4.81VVICDD223 pKa = 4.01DD224 pKa = 3.29VDD226 pKa = 3.35EE227 pKa = 4.54RR228 pKa = 11.84TVPEE232 pKa = 3.86QQEE235 pKa = 4.01IKK237 pKa = 8.79TWCDD241 pKa = 3.15LDD243 pKa = 3.87PFQVKK248 pKa = 9.35VNYY251 pKa = 9.43KK252 pKa = 9.77VLNIRR257 pKa = 11.84PKK259 pKa = 10.71RR260 pKa = 11.84FIFTSNEE267 pKa = 4.0SIADD271 pKa = 3.68CFPRR275 pKa = 11.84AKK277 pKa = 10.05PIHH280 pKa = 6.25LKK282 pKa = 10.44AIEE285 pKa = 4.01RR286 pKa = 11.84RR287 pKa = 11.84FKK289 pKa = 10.78VYY291 pKa = 10.23YY292 pKa = 9.83YY293 pKa = 9.07PAAYY297 pKa = 9.77GEE299 pKa = 4.66PGWVDD304 pKa = 3.39PTAIPQKK311 pKa = 10.94YY312 pKa = 9.36LFTDD316 pKa = 3.66EE317 pKa = 5.52EE318 pKa = 4.1MDD320 pKa = 3.48AQLSVV325 pKa = 3.14
MM1 pKa = 7.53PPKK4 pKa = 10.4AQFNRR9 pKa = 11.84FCQITKK15 pKa = 10.26KK16 pKa = 10.52RR17 pKa = 11.84GFCWTYY23 pKa = 10.59HH24 pKa = 7.02DD25 pKa = 4.68YY26 pKa = 11.56DD27 pKa = 4.73DD28 pKa = 6.15EE29 pKa = 4.82IVAHH33 pKa = 6.61IKK35 pKa = 10.92GLFDD39 pKa = 5.37RR40 pKa = 11.84PNGWVKK46 pKa = 10.57RR47 pKa = 11.84LVFNYY52 pKa = 7.44EE53 pKa = 3.54ICPTTGRR60 pKa = 11.84RR61 pKa = 11.84HH62 pKa = 5.42LQGCLYY68 pKa = 10.25MRR70 pKa = 11.84DD71 pKa = 3.49NQTWPKK77 pKa = 10.14LKK79 pKa = 10.61ADD81 pKa = 3.41IGLEE85 pKa = 4.18KK86 pKa = 10.33IHH88 pKa = 7.17LEE90 pKa = 3.72LAQNVEE96 pKa = 4.71ALDD99 pKa = 4.26KK100 pKa = 10.78YY101 pKa = 9.03CQKK104 pKa = 10.87SEE106 pKa = 4.19TRR108 pKa = 11.84DD109 pKa = 3.36PAHH112 pKa = 7.13PEE114 pKa = 3.99GVVLGVAPLTDD125 pKa = 3.34EE126 pKa = 5.21AKK128 pKa = 9.22GQKK131 pKa = 10.11GEE133 pKa = 4.06EE134 pKa = 4.3YY135 pKa = 10.63YY136 pKa = 10.81EE137 pKa = 4.38RR138 pKa = 11.84NILACMNGQQMDD150 pKa = 3.56ASAEE154 pKa = 3.97FNLRR158 pKa = 11.84NFEE161 pKa = 4.15YY162 pKa = 10.54AAEE165 pKa = 3.9ARR167 pKa = 11.84KK168 pKa = 9.6RR169 pKa = 11.84KK170 pKa = 9.86RR171 pKa = 11.84EE172 pKa = 3.92LASLDD177 pKa = 3.5EE178 pKa = 5.91LSFEE182 pKa = 4.01WHH184 pKa = 6.01YY185 pKa = 11.75GEE187 pKa = 4.75PFSGKK192 pKa = 6.61THH194 pKa = 5.96YY195 pKa = 10.01CRR197 pKa = 11.84KK198 pKa = 9.73IPGAFKK204 pKa = 10.13WNSKK208 pKa = 10.48AGWNNYY214 pKa = 8.25NDD216 pKa = 4.18EE217 pKa = 4.43EE218 pKa = 4.81VVICDD223 pKa = 4.01DD224 pKa = 3.29VDD226 pKa = 3.35EE227 pKa = 4.54RR228 pKa = 11.84TVPEE232 pKa = 3.86QQEE235 pKa = 4.01IKK237 pKa = 8.79TWCDD241 pKa = 3.15LDD243 pKa = 3.87PFQVKK248 pKa = 9.35VNYY251 pKa = 9.43KK252 pKa = 9.77VLNIRR257 pKa = 11.84PKK259 pKa = 10.71RR260 pKa = 11.84FIFTSNEE267 pKa = 4.0SIADD271 pKa = 3.68CFPRR275 pKa = 11.84AKK277 pKa = 10.05PIHH280 pKa = 6.25LKK282 pKa = 10.44AIEE285 pKa = 4.01RR286 pKa = 11.84RR287 pKa = 11.84FKK289 pKa = 10.78VYY291 pKa = 10.23YY292 pKa = 9.83YY293 pKa = 9.07PAAYY297 pKa = 9.77GEE299 pKa = 4.66PGWVDD304 pKa = 3.39PTAIPQKK311 pKa = 10.94YY312 pKa = 9.36LFTDD316 pKa = 3.66EE317 pKa = 5.52EE318 pKa = 4.1MDD320 pKa = 3.48AQLSVV325 pKa = 3.14
Molecular weight: 38.02 kDa
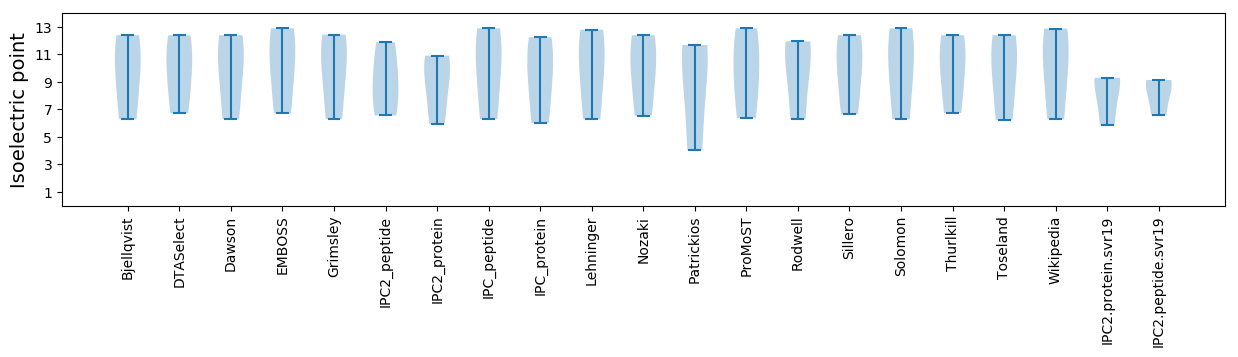
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A190WHM0|A0A190WHM0_9CIRC Uncharacterized protein OS=Circovirus-like genome DCCV-12 OX=1788440 PE=4 SV=1
MM1 pKa = 7.38ASKK4 pKa = 7.96WTPARR9 pKa = 11.84SSIYY13 pKa = 9.0GTSSTLPRR21 pKa = 11.84LANANANLLHH31 pKa = 6.58SMSFHH36 pKa = 6.9SSGITASLLAARR48 pKa = 11.84LIIAVRR54 pKa = 11.84SQVPSNGIRR63 pKa = 11.84RR64 pKa = 11.84LVGTTTTMRR73 pKa = 11.84RR74 pKa = 11.84SSYY77 pKa = 11.29VMTSMNVLFPSNRR90 pKa = 11.84RR91 pKa = 11.84LRR93 pKa = 11.84PGVISIPSRR102 pKa = 11.84SRR104 pKa = 11.84LITRR108 pKa = 11.84CC109 pKa = 3.32
MM1 pKa = 7.38ASKK4 pKa = 7.96WTPARR9 pKa = 11.84SSIYY13 pKa = 9.0GTSSTLPRR21 pKa = 11.84LANANANLLHH31 pKa = 6.58SMSFHH36 pKa = 6.9SSGITASLLAARR48 pKa = 11.84LIIAVRR54 pKa = 11.84SQVPSNGIRR63 pKa = 11.84RR64 pKa = 11.84LVGTTTTMRR73 pKa = 11.84RR74 pKa = 11.84SSYY77 pKa = 11.29VMTSMNVLFPSNRR90 pKa = 11.84RR91 pKa = 11.84LRR93 pKa = 11.84PGVISIPSRR102 pKa = 11.84SRR104 pKa = 11.84LITRR108 pKa = 11.84CC109 pKa = 3.32
Molecular weight: 11.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
727 |
109 |
325 |
242.3 |
27.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.978 ± 0.362 | 1.513 ± 0.962 |
4.677 ± 1.542 | 4.952 ± 2.334 |
4.539 ± 0.826 | 5.365 ± 0.298 |
1.788 ± 0.444 | 5.502 ± 0.58 |
7.153 ± 1.907 | 7.153 ± 0.899 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.751 ± 0.795 | 5.915 ± 0.705 |
6.052 ± 0.26 | 3.301 ± 0.812 |
7.703 ± 1.61 | 5.777 ± 3.586 |
5.365 ± 1.235 | 6.327 ± 0.85 |
1.651 ± 0.492 | 4.539 ± 0.832 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
