
Beihai narna-like virus 15
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
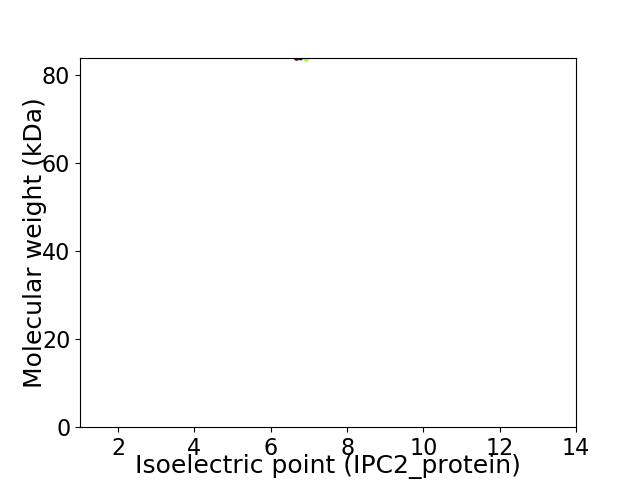
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KHW5|A0A1L3KHW5_9VIRU RNA-dependent RNA polymerase OS=Beihai narna-like virus 15 OX=1922442 PE=4 SV=1
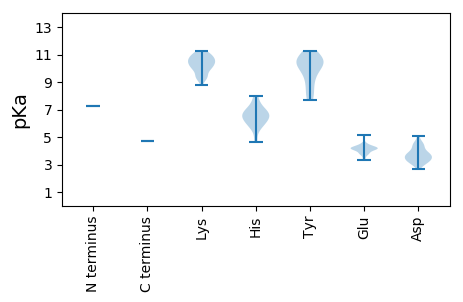
MM1 pKa = 7.22QSKK4 pKa = 10.28ALAATVPEE12 pKa = 4.27SFILEE17 pKa = 4.22SLLKK21 pKa = 10.57HH22 pKa = 6.41CEE24 pKa = 4.06TVSKK28 pKa = 9.39VTQTSKK34 pKa = 11.08EE35 pKa = 3.8LLEE38 pKa = 4.25EE39 pKa = 4.11FKK41 pKa = 10.67EE42 pKa = 4.14YY43 pKa = 10.41CRR45 pKa = 11.84PIMRR49 pKa = 11.84DD50 pKa = 2.7AAQRR54 pKa = 11.84LSSTYY59 pKa = 10.59SGIPSNHH66 pKa = 6.33ASYY69 pKa = 8.02EE70 pKa = 4.14TTRR73 pKa = 11.84EE74 pKa = 3.79KK75 pKa = 11.22GGVGRR80 pKa = 11.84NYY82 pKa = 10.54KK83 pKa = 10.3LKK85 pKa = 10.71RR86 pKa = 11.84NLFFQPEE93 pKa = 4.2NPRR96 pKa = 11.84LDD98 pKa = 5.09PITTLISGKK107 pKa = 10.52AGIGKK112 pKa = 9.36SLLQAKK118 pKa = 9.26IVSKK122 pKa = 10.16MADD125 pKa = 3.17VLGRR129 pKa = 11.84SPFQTSYY136 pKa = 11.09NRR138 pKa = 11.84NSDD141 pKa = 3.59TEE143 pKa = 3.83HH144 pKa = 6.43WDD146 pKa = 3.69GYY148 pKa = 7.83NQQPIVMIDD157 pKa = 3.54DD158 pKa = 4.59FQQLRR163 pKa = 11.84TRR165 pKa = 11.84NYY167 pKa = 10.7DD168 pKa = 3.23VQQEE172 pKa = 4.19EE173 pKa = 5.12KK174 pKa = 11.09EE175 pKa = 4.4FITLNSTVDD184 pKa = 3.12KK185 pKa = 9.68VLPMAHH191 pKa = 6.79LKK193 pKa = 10.79DD194 pKa = 3.44KK195 pKa = 10.82GMKK198 pKa = 9.67FNSPFIIYY206 pKa = 10.21SSNHH210 pKa = 4.65TLGEE214 pKa = 4.54TICEE218 pKa = 4.16LQRR221 pKa = 11.84NFHH224 pKa = 5.83SFRR227 pKa = 11.84AIEE230 pKa = 3.79RR231 pKa = 11.84RR232 pKa = 11.84FDD234 pKa = 3.45YY235 pKa = 11.26LIDD238 pKa = 4.12YY239 pKa = 8.91DD240 pKa = 4.34PKK242 pKa = 11.22GIGYY246 pKa = 9.37WMVKK250 pKa = 9.94KK251 pKa = 10.05VVPYY255 pKa = 10.22QYY257 pKa = 10.83SDD259 pKa = 3.24SQRR262 pKa = 11.84QTNPFMVKK270 pKa = 9.37MEE272 pKa = 3.95QMPLTPNLDD281 pKa = 2.96RR282 pKa = 11.84HH283 pKa = 5.79FQRR286 pKa = 11.84QTIFGSSSIDD296 pKa = 3.48EE297 pKa = 3.99IASFFTHH304 pKa = 6.93LMIKK308 pKa = 8.78EE309 pKa = 4.21WEE311 pKa = 4.27VKK313 pKa = 10.15IIFYY317 pKa = 10.62QNEE320 pKa = 3.71IRR322 pKa = 11.84NLNLVPIGPGIHH334 pKa = 6.54GEE336 pKa = 4.02WISNEE341 pKa = 3.43KK342 pKa = 9.3HH343 pKa = 6.28NKK345 pKa = 9.25VKK347 pKa = 10.59VKK349 pKa = 10.46PILEE353 pKa = 4.14PLKK356 pKa = 10.92VRR358 pKa = 11.84TITIGSSQNFMLKK371 pKa = 10.02PISKK375 pKa = 10.51AIFQSLQKK383 pKa = 9.92YY384 pKa = 10.18PEE386 pKa = 4.24MKK388 pKa = 9.95PCFTPEE394 pKa = 4.16YY395 pKa = 8.39IDD397 pKa = 3.38SVEE400 pKa = 4.12RR401 pKa = 11.84LKK403 pKa = 11.03ALPGLWLSGDD413 pKa = 3.7YY414 pKa = 11.06SSATDD419 pKa = 4.12GLHH422 pKa = 6.89SDD424 pKa = 4.47LFRR427 pKa = 11.84AGLSTLEE434 pKa = 4.36EE435 pKa = 4.27YY436 pKa = 10.74LPCEE440 pKa = 3.44LWKK443 pKa = 10.55LVKK446 pKa = 10.27RR447 pKa = 11.84EE448 pKa = 4.14MDD450 pKa = 3.33PHH452 pKa = 7.47LCEE455 pKa = 4.06YY456 pKa = 10.48PEE458 pKa = 4.36GLGIPDD464 pKa = 3.79VFQTNGQLMGSLLSFPLLSLANAFTLSKK492 pKa = 9.95TLDD495 pKa = 3.65RR496 pKa = 11.84PLGDD500 pKa = 4.54LPCLIHH506 pKa = 7.96GDD508 pKa = 3.75DD509 pKa = 4.5LLAKK513 pKa = 10.42VSLDD517 pKa = 4.32QYY519 pKa = 11.1LDD521 pKa = 3.15WKK523 pKa = 10.66AFCPKK528 pKa = 9.88IGLSLSVGKK537 pKa = 10.41NYY539 pKa = 9.78ISEE542 pKa = 4.1HH543 pKa = 5.85WGSIDD548 pKa = 3.32SQVYY552 pKa = 9.46FEE554 pKa = 4.52SEE556 pKa = 3.78KK557 pKa = 11.15LGTGKK562 pKa = 10.33FSALFSKK569 pKa = 10.64EE570 pKa = 3.84ITCLPTLVRR579 pKa = 11.84RR580 pKa = 11.84GVPKK584 pKa = 10.2PLIVSLLKK592 pKa = 10.63KK593 pKa = 10.08EE594 pKa = 4.23IEE596 pKa = 4.07RR597 pKa = 11.84TPRR600 pKa = 11.84SLDD603 pKa = 3.14VSQEE607 pKa = 3.79FGGLGIHH614 pKa = 6.63GEE616 pKa = 4.37PYY618 pKa = 10.27DD619 pKa = 3.75SRR621 pKa = 11.84SRR623 pKa = 11.84AVFHH627 pKa = 6.93NMVLKK632 pKa = 10.88SFGKK636 pKa = 9.26RR637 pKa = 11.84EE638 pKa = 3.69VWGGYY643 pKa = 9.27IYY645 pKa = 10.31TISEE649 pKa = 4.33SLKK652 pKa = 10.89DD653 pKa = 3.84MVPQRR658 pKa = 11.84YY659 pKa = 8.59QSPIEE664 pKa = 4.23IPDD667 pKa = 3.84DD668 pKa = 3.9LEE670 pKa = 4.39VYY672 pKa = 10.05KK673 pKa = 10.97KK674 pKa = 10.94NEE676 pKa = 3.32FRR678 pKa = 11.84EE679 pKa = 4.15IKK681 pKa = 10.32RR682 pKa = 11.84FEE684 pKa = 4.27NLTFSQNYY692 pKa = 7.67TPITKK697 pKa = 9.53WSTFSEE703 pKa = 4.58SEE705 pKa = 4.12NPFLEE710 pKa = 4.08NLISQSRR717 pKa = 11.84NNYY720 pKa = 9.97SEE722 pKa = 4.22VLTTFDD728 pKa = 3.57FF729 pKa = 4.72
MM1 pKa = 7.22QSKK4 pKa = 10.28ALAATVPEE12 pKa = 4.27SFILEE17 pKa = 4.22SLLKK21 pKa = 10.57HH22 pKa = 6.41CEE24 pKa = 4.06TVSKK28 pKa = 9.39VTQTSKK34 pKa = 11.08EE35 pKa = 3.8LLEE38 pKa = 4.25EE39 pKa = 4.11FKK41 pKa = 10.67EE42 pKa = 4.14YY43 pKa = 10.41CRR45 pKa = 11.84PIMRR49 pKa = 11.84DD50 pKa = 2.7AAQRR54 pKa = 11.84LSSTYY59 pKa = 10.59SGIPSNHH66 pKa = 6.33ASYY69 pKa = 8.02EE70 pKa = 4.14TTRR73 pKa = 11.84EE74 pKa = 3.79KK75 pKa = 11.22GGVGRR80 pKa = 11.84NYY82 pKa = 10.54KK83 pKa = 10.3LKK85 pKa = 10.71RR86 pKa = 11.84NLFFQPEE93 pKa = 4.2NPRR96 pKa = 11.84LDD98 pKa = 5.09PITTLISGKK107 pKa = 10.52AGIGKK112 pKa = 9.36SLLQAKK118 pKa = 9.26IVSKK122 pKa = 10.16MADD125 pKa = 3.17VLGRR129 pKa = 11.84SPFQTSYY136 pKa = 11.09NRR138 pKa = 11.84NSDD141 pKa = 3.59TEE143 pKa = 3.83HH144 pKa = 6.43WDD146 pKa = 3.69GYY148 pKa = 7.83NQQPIVMIDD157 pKa = 3.54DD158 pKa = 4.59FQQLRR163 pKa = 11.84TRR165 pKa = 11.84NYY167 pKa = 10.7DD168 pKa = 3.23VQQEE172 pKa = 4.19EE173 pKa = 5.12KK174 pKa = 11.09EE175 pKa = 4.4FITLNSTVDD184 pKa = 3.12KK185 pKa = 9.68VLPMAHH191 pKa = 6.79LKK193 pKa = 10.79DD194 pKa = 3.44KK195 pKa = 10.82GMKK198 pKa = 9.67FNSPFIIYY206 pKa = 10.21SSNHH210 pKa = 4.65TLGEE214 pKa = 4.54TICEE218 pKa = 4.16LQRR221 pKa = 11.84NFHH224 pKa = 5.83SFRR227 pKa = 11.84AIEE230 pKa = 3.79RR231 pKa = 11.84RR232 pKa = 11.84FDD234 pKa = 3.45YY235 pKa = 11.26LIDD238 pKa = 4.12YY239 pKa = 8.91DD240 pKa = 4.34PKK242 pKa = 11.22GIGYY246 pKa = 9.37WMVKK250 pKa = 9.94KK251 pKa = 10.05VVPYY255 pKa = 10.22QYY257 pKa = 10.83SDD259 pKa = 3.24SQRR262 pKa = 11.84QTNPFMVKK270 pKa = 9.37MEE272 pKa = 3.95QMPLTPNLDD281 pKa = 2.96RR282 pKa = 11.84HH283 pKa = 5.79FQRR286 pKa = 11.84QTIFGSSSIDD296 pKa = 3.48EE297 pKa = 3.99IASFFTHH304 pKa = 6.93LMIKK308 pKa = 8.78EE309 pKa = 4.21WEE311 pKa = 4.27VKK313 pKa = 10.15IIFYY317 pKa = 10.62QNEE320 pKa = 3.71IRR322 pKa = 11.84NLNLVPIGPGIHH334 pKa = 6.54GEE336 pKa = 4.02WISNEE341 pKa = 3.43KK342 pKa = 9.3HH343 pKa = 6.28NKK345 pKa = 9.25VKK347 pKa = 10.59VKK349 pKa = 10.46PILEE353 pKa = 4.14PLKK356 pKa = 10.92VRR358 pKa = 11.84TITIGSSQNFMLKK371 pKa = 10.02PISKK375 pKa = 10.51AIFQSLQKK383 pKa = 9.92YY384 pKa = 10.18PEE386 pKa = 4.24MKK388 pKa = 9.95PCFTPEE394 pKa = 4.16YY395 pKa = 8.39IDD397 pKa = 3.38SVEE400 pKa = 4.12RR401 pKa = 11.84LKK403 pKa = 11.03ALPGLWLSGDD413 pKa = 3.7YY414 pKa = 11.06SSATDD419 pKa = 4.12GLHH422 pKa = 6.89SDD424 pKa = 4.47LFRR427 pKa = 11.84AGLSTLEE434 pKa = 4.36EE435 pKa = 4.27YY436 pKa = 10.74LPCEE440 pKa = 3.44LWKK443 pKa = 10.55LVKK446 pKa = 10.27RR447 pKa = 11.84EE448 pKa = 4.14MDD450 pKa = 3.33PHH452 pKa = 7.47LCEE455 pKa = 4.06YY456 pKa = 10.48PEE458 pKa = 4.36GLGIPDD464 pKa = 3.79VFQTNGQLMGSLLSFPLLSLANAFTLSKK492 pKa = 9.95TLDD495 pKa = 3.65RR496 pKa = 11.84PLGDD500 pKa = 4.54LPCLIHH506 pKa = 7.96GDD508 pKa = 3.75DD509 pKa = 4.5LLAKK513 pKa = 10.42VSLDD517 pKa = 4.32QYY519 pKa = 11.1LDD521 pKa = 3.15WKK523 pKa = 10.66AFCPKK528 pKa = 9.88IGLSLSVGKK537 pKa = 10.41NYY539 pKa = 9.78ISEE542 pKa = 4.1HH543 pKa = 5.85WGSIDD548 pKa = 3.32SQVYY552 pKa = 9.46FEE554 pKa = 4.52SEE556 pKa = 3.78KK557 pKa = 11.15LGTGKK562 pKa = 10.33FSALFSKK569 pKa = 10.64EE570 pKa = 3.84ITCLPTLVRR579 pKa = 11.84RR580 pKa = 11.84GVPKK584 pKa = 10.2PLIVSLLKK592 pKa = 10.63KK593 pKa = 10.08EE594 pKa = 4.23IEE596 pKa = 4.07RR597 pKa = 11.84TPRR600 pKa = 11.84SLDD603 pKa = 3.14VSQEE607 pKa = 3.79FGGLGIHH614 pKa = 6.63GEE616 pKa = 4.37PYY618 pKa = 10.27DD619 pKa = 3.75SRR621 pKa = 11.84SRR623 pKa = 11.84AVFHH627 pKa = 6.93NMVLKK632 pKa = 10.88SFGKK636 pKa = 9.26RR637 pKa = 11.84EE638 pKa = 3.69VWGGYY643 pKa = 9.27IYY645 pKa = 10.31TISEE649 pKa = 4.33SLKK652 pKa = 10.89DD653 pKa = 3.84MVPQRR658 pKa = 11.84YY659 pKa = 8.59QSPIEE664 pKa = 4.23IPDD667 pKa = 3.84DD668 pKa = 3.9LEE670 pKa = 4.39VYY672 pKa = 10.05KK673 pKa = 10.97KK674 pKa = 10.94NEE676 pKa = 3.32FRR678 pKa = 11.84EE679 pKa = 4.15IKK681 pKa = 10.32RR682 pKa = 11.84FEE684 pKa = 4.27NLTFSQNYY692 pKa = 7.67TPITKK697 pKa = 9.53WSTFSEE703 pKa = 4.58SEE705 pKa = 4.12NPFLEE710 pKa = 4.08NLISQSRR717 pKa = 11.84NNYY720 pKa = 9.97SEE722 pKa = 4.22VLTTFDD728 pKa = 3.57FF729 pKa = 4.72
Molecular weight: 83.73 kDa
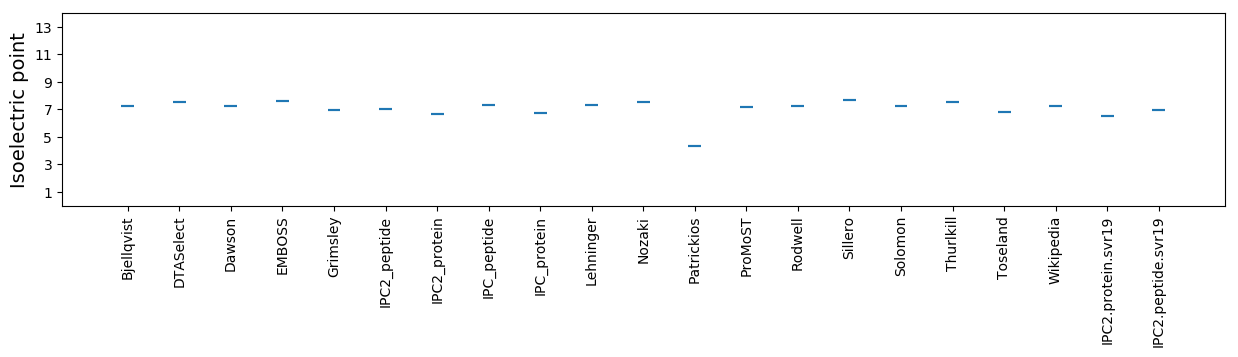
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KHW5|A0A1L3KHW5_9VIRU RNA-dependent RNA polymerase OS=Beihai narna-like virus 15 OX=1922442 PE=4 SV=1
MM1 pKa = 7.22QSKK4 pKa = 10.28ALAATVPEE12 pKa = 4.27SFILEE17 pKa = 4.22SLLKK21 pKa = 10.57HH22 pKa = 6.41CEE24 pKa = 4.06TVSKK28 pKa = 9.39VTQTSKK34 pKa = 11.08EE35 pKa = 3.8LLEE38 pKa = 4.25EE39 pKa = 4.11FKK41 pKa = 10.67EE42 pKa = 4.14YY43 pKa = 10.41CRR45 pKa = 11.84PIMRR49 pKa = 11.84DD50 pKa = 2.7AAQRR54 pKa = 11.84LSSTYY59 pKa = 10.59SGIPSNHH66 pKa = 6.33ASYY69 pKa = 8.02EE70 pKa = 4.14TTRR73 pKa = 11.84EE74 pKa = 3.79KK75 pKa = 11.22GGVGRR80 pKa = 11.84NYY82 pKa = 10.54KK83 pKa = 10.3LKK85 pKa = 10.71RR86 pKa = 11.84NLFFQPEE93 pKa = 4.2NPRR96 pKa = 11.84LDD98 pKa = 5.09PITTLISGKK107 pKa = 10.52AGIGKK112 pKa = 9.36SLLQAKK118 pKa = 9.26IVSKK122 pKa = 10.16MADD125 pKa = 3.17VLGRR129 pKa = 11.84SPFQTSYY136 pKa = 11.09NRR138 pKa = 11.84NSDD141 pKa = 3.59TEE143 pKa = 3.83HH144 pKa = 6.43WDD146 pKa = 3.69GYY148 pKa = 7.83NQQPIVMIDD157 pKa = 3.54DD158 pKa = 4.59FQQLRR163 pKa = 11.84TRR165 pKa = 11.84NYY167 pKa = 10.7DD168 pKa = 3.23VQQEE172 pKa = 4.19EE173 pKa = 5.12KK174 pKa = 11.09EE175 pKa = 4.4FITLNSTVDD184 pKa = 3.12KK185 pKa = 9.68VLPMAHH191 pKa = 6.79LKK193 pKa = 10.79DD194 pKa = 3.44KK195 pKa = 10.82GMKK198 pKa = 9.67FNSPFIIYY206 pKa = 10.21SSNHH210 pKa = 4.65TLGEE214 pKa = 4.54TICEE218 pKa = 4.16LQRR221 pKa = 11.84NFHH224 pKa = 5.83SFRR227 pKa = 11.84AIEE230 pKa = 3.79RR231 pKa = 11.84RR232 pKa = 11.84FDD234 pKa = 3.45YY235 pKa = 11.26LIDD238 pKa = 4.12YY239 pKa = 8.91DD240 pKa = 4.34PKK242 pKa = 11.22GIGYY246 pKa = 9.37WMVKK250 pKa = 9.94KK251 pKa = 10.05VVPYY255 pKa = 10.22QYY257 pKa = 10.83SDD259 pKa = 3.24SQRR262 pKa = 11.84QTNPFMVKK270 pKa = 9.37MEE272 pKa = 3.95QMPLTPNLDD281 pKa = 2.96RR282 pKa = 11.84HH283 pKa = 5.79FQRR286 pKa = 11.84QTIFGSSSIDD296 pKa = 3.48EE297 pKa = 3.99IASFFTHH304 pKa = 6.93LMIKK308 pKa = 8.78EE309 pKa = 4.21WEE311 pKa = 4.27VKK313 pKa = 10.15IIFYY317 pKa = 10.62QNEE320 pKa = 3.71IRR322 pKa = 11.84NLNLVPIGPGIHH334 pKa = 6.54GEE336 pKa = 4.02WISNEE341 pKa = 3.43KK342 pKa = 9.3HH343 pKa = 6.28NKK345 pKa = 9.25VKK347 pKa = 10.59VKK349 pKa = 10.46PILEE353 pKa = 4.14PLKK356 pKa = 10.92VRR358 pKa = 11.84TITIGSSQNFMLKK371 pKa = 10.02PISKK375 pKa = 10.51AIFQSLQKK383 pKa = 9.92YY384 pKa = 10.18PEE386 pKa = 4.24MKK388 pKa = 9.95PCFTPEE394 pKa = 4.16YY395 pKa = 8.39IDD397 pKa = 3.38SVEE400 pKa = 4.12RR401 pKa = 11.84LKK403 pKa = 11.03ALPGLWLSGDD413 pKa = 3.7YY414 pKa = 11.06SSATDD419 pKa = 4.12GLHH422 pKa = 6.89SDD424 pKa = 4.47LFRR427 pKa = 11.84AGLSTLEE434 pKa = 4.36EE435 pKa = 4.27YY436 pKa = 10.74LPCEE440 pKa = 3.44LWKK443 pKa = 10.55LVKK446 pKa = 10.27RR447 pKa = 11.84EE448 pKa = 4.14MDD450 pKa = 3.33PHH452 pKa = 7.47LCEE455 pKa = 4.06YY456 pKa = 10.48PEE458 pKa = 4.36GLGIPDD464 pKa = 3.79VFQTNGQLMGSLLSFPLLSLANAFTLSKK492 pKa = 9.95TLDD495 pKa = 3.65RR496 pKa = 11.84PLGDD500 pKa = 4.54LPCLIHH506 pKa = 7.96GDD508 pKa = 3.75DD509 pKa = 4.5LLAKK513 pKa = 10.42VSLDD517 pKa = 4.32QYY519 pKa = 11.1LDD521 pKa = 3.15WKK523 pKa = 10.66AFCPKK528 pKa = 9.88IGLSLSVGKK537 pKa = 10.41NYY539 pKa = 9.78ISEE542 pKa = 4.1HH543 pKa = 5.85WGSIDD548 pKa = 3.32SQVYY552 pKa = 9.46FEE554 pKa = 4.52SEE556 pKa = 3.78KK557 pKa = 11.15LGTGKK562 pKa = 10.33FSALFSKK569 pKa = 10.64EE570 pKa = 3.84ITCLPTLVRR579 pKa = 11.84RR580 pKa = 11.84GVPKK584 pKa = 10.2PLIVSLLKK592 pKa = 10.63KK593 pKa = 10.08EE594 pKa = 4.23IEE596 pKa = 4.07RR597 pKa = 11.84TPRR600 pKa = 11.84SLDD603 pKa = 3.14VSQEE607 pKa = 3.79FGGLGIHH614 pKa = 6.63GEE616 pKa = 4.37PYY618 pKa = 10.27DD619 pKa = 3.75SRR621 pKa = 11.84SRR623 pKa = 11.84AVFHH627 pKa = 6.93NMVLKK632 pKa = 10.88SFGKK636 pKa = 9.26RR637 pKa = 11.84EE638 pKa = 3.69VWGGYY643 pKa = 9.27IYY645 pKa = 10.31TISEE649 pKa = 4.33SLKK652 pKa = 10.89DD653 pKa = 3.84MVPQRR658 pKa = 11.84YY659 pKa = 8.59QSPIEE664 pKa = 4.23IPDD667 pKa = 3.84DD668 pKa = 3.9LEE670 pKa = 4.39VYY672 pKa = 10.05KK673 pKa = 10.97KK674 pKa = 10.94NEE676 pKa = 3.32FRR678 pKa = 11.84EE679 pKa = 4.15IKK681 pKa = 10.32RR682 pKa = 11.84FEE684 pKa = 4.27NLTFSQNYY692 pKa = 7.67TPITKK697 pKa = 9.53WSTFSEE703 pKa = 4.58SEE705 pKa = 4.12NPFLEE710 pKa = 4.08NLISQSRR717 pKa = 11.84NNYY720 pKa = 9.97SEE722 pKa = 4.22VLTTFDD728 pKa = 3.57FF729 pKa = 4.72
MM1 pKa = 7.22QSKK4 pKa = 10.28ALAATVPEE12 pKa = 4.27SFILEE17 pKa = 4.22SLLKK21 pKa = 10.57HH22 pKa = 6.41CEE24 pKa = 4.06TVSKK28 pKa = 9.39VTQTSKK34 pKa = 11.08EE35 pKa = 3.8LLEE38 pKa = 4.25EE39 pKa = 4.11FKK41 pKa = 10.67EE42 pKa = 4.14YY43 pKa = 10.41CRR45 pKa = 11.84PIMRR49 pKa = 11.84DD50 pKa = 2.7AAQRR54 pKa = 11.84LSSTYY59 pKa = 10.59SGIPSNHH66 pKa = 6.33ASYY69 pKa = 8.02EE70 pKa = 4.14TTRR73 pKa = 11.84EE74 pKa = 3.79KK75 pKa = 11.22GGVGRR80 pKa = 11.84NYY82 pKa = 10.54KK83 pKa = 10.3LKK85 pKa = 10.71RR86 pKa = 11.84NLFFQPEE93 pKa = 4.2NPRR96 pKa = 11.84LDD98 pKa = 5.09PITTLISGKK107 pKa = 10.52AGIGKK112 pKa = 9.36SLLQAKK118 pKa = 9.26IVSKK122 pKa = 10.16MADD125 pKa = 3.17VLGRR129 pKa = 11.84SPFQTSYY136 pKa = 11.09NRR138 pKa = 11.84NSDD141 pKa = 3.59TEE143 pKa = 3.83HH144 pKa = 6.43WDD146 pKa = 3.69GYY148 pKa = 7.83NQQPIVMIDD157 pKa = 3.54DD158 pKa = 4.59FQQLRR163 pKa = 11.84TRR165 pKa = 11.84NYY167 pKa = 10.7DD168 pKa = 3.23VQQEE172 pKa = 4.19EE173 pKa = 5.12KK174 pKa = 11.09EE175 pKa = 4.4FITLNSTVDD184 pKa = 3.12KK185 pKa = 9.68VLPMAHH191 pKa = 6.79LKK193 pKa = 10.79DD194 pKa = 3.44KK195 pKa = 10.82GMKK198 pKa = 9.67FNSPFIIYY206 pKa = 10.21SSNHH210 pKa = 4.65TLGEE214 pKa = 4.54TICEE218 pKa = 4.16LQRR221 pKa = 11.84NFHH224 pKa = 5.83SFRR227 pKa = 11.84AIEE230 pKa = 3.79RR231 pKa = 11.84RR232 pKa = 11.84FDD234 pKa = 3.45YY235 pKa = 11.26LIDD238 pKa = 4.12YY239 pKa = 8.91DD240 pKa = 4.34PKK242 pKa = 11.22GIGYY246 pKa = 9.37WMVKK250 pKa = 9.94KK251 pKa = 10.05VVPYY255 pKa = 10.22QYY257 pKa = 10.83SDD259 pKa = 3.24SQRR262 pKa = 11.84QTNPFMVKK270 pKa = 9.37MEE272 pKa = 3.95QMPLTPNLDD281 pKa = 2.96RR282 pKa = 11.84HH283 pKa = 5.79FQRR286 pKa = 11.84QTIFGSSSIDD296 pKa = 3.48EE297 pKa = 3.99IASFFTHH304 pKa = 6.93LMIKK308 pKa = 8.78EE309 pKa = 4.21WEE311 pKa = 4.27VKK313 pKa = 10.15IIFYY317 pKa = 10.62QNEE320 pKa = 3.71IRR322 pKa = 11.84NLNLVPIGPGIHH334 pKa = 6.54GEE336 pKa = 4.02WISNEE341 pKa = 3.43KK342 pKa = 9.3HH343 pKa = 6.28NKK345 pKa = 9.25VKK347 pKa = 10.59VKK349 pKa = 10.46PILEE353 pKa = 4.14PLKK356 pKa = 10.92VRR358 pKa = 11.84TITIGSSQNFMLKK371 pKa = 10.02PISKK375 pKa = 10.51AIFQSLQKK383 pKa = 9.92YY384 pKa = 10.18PEE386 pKa = 4.24MKK388 pKa = 9.95PCFTPEE394 pKa = 4.16YY395 pKa = 8.39IDD397 pKa = 3.38SVEE400 pKa = 4.12RR401 pKa = 11.84LKK403 pKa = 11.03ALPGLWLSGDD413 pKa = 3.7YY414 pKa = 11.06SSATDD419 pKa = 4.12GLHH422 pKa = 6.89SDD424 pKa = 4.47LFRR427 pKa = 11.84AGLSTLEE434 pKa = 4.36EE435 pKa = 4.27YY436 pKa = 10.74LPCEE440 pKa = 3.44LWKK443 pKa = 10.55LVKK446 pKa = 10.27RR447 pKa = 11.84EE448 pKa = 4.14MDD450 pKa = 3.33PHH452 pKa = 7.47LCEE455 pKa = 4.06YY456 pKa = 10.48PEE458 pKa = 4.36GLGIPDD464 pKa = 3.79VFQTNGQLMGSLLSFPLLSLANAFTLSKK492 pKa = 9.95TLDD495 pKa = 3.65RR496 pKa = 11.84PLGDD500 pKa = 4.54LPCLIHH506 pKa = 7.96GDD508 pKa = 3.75DD509 pKa = 4.5LLAKK513 pKa = 10.42VSLDD517 pKa = 4.32QYY519 pKa = 11.1LDD521 pKa = 3.15WKK523 pKa = 10.66AFCPKK528 pKa = 9.88IGLSLSVGKK537 pKa = 10.41NYY539 pKa = 9.78ISEE542 pKa = 4.1HH543 pKa = 5.85WGSIDD548 pKa = 3.32SQVYY552 pKa = 9.46FEE554 pKa = 4.52SEE556 pKa = 3.78KK557 pKa = 11.15LGTGKK562 pKa = 10.33FSALFSKK569 pKa = 10.64EE570 pKa = 3.84ITCLPTLVRR579 pKa = 11.84RR580 pKa = 11.84GVPKK584 pKa = 10.2PLIVSLLKK592 pKa = 10.63KK593 pKa = 10.08EE594 pKa = 4.23IEE596 pKa = 4.07RR597 pKa = 11.84TPRR600 pKa = 11.84SLDD603 pKa = 3.14VSQEE607 pKa = 3.79FGGLGIHH614 pKa = 6.63GEE616 pKa = 4.37PYY618 pKa = 10.27DD619 pKa = 3.75SRR621 pKa = 11.84SRR623 pKa = 11.84AVFHH627 pKa = 6.93NMVLKK632 pKa = 10.88SFGKK636 pKa = 9.26RR637 pKa = 11.84EE638 pKa = 3.69VWGGYY643 pKa = 9.27IYY645 pKa = 10.31TISEE649 pKa = 4.33SLKK652 pKa = 10.89DD653 pKa = 3.84MVPQRR658 pKa = 11.84YY659 pKa = 8.59QSPIEE664 pKa = 4.23IPDD667 pKa = 3.84DD668 pKa = 3.9LEE670 pKa = 4.39VYY672 pKa = 10.05KK673 pKa = 10.97KK674 pKa = 10.94NEE676 pKa = 3.32FRR678 pKa = 11.84EE679 pKa = 4.15IKK681 pKa = 10.32RR682 pKa = 11.84FEE684 pKa = 4.27NLTFSQNYY692 pKa = 7.67TPITKK697 pKa = 9.53WSTFSEE703 pKa = 4.58SEE705 pKa = 4.12NPFLEE710 pKa = 4.08NLISQSRR717 pKa = 11.84NNYY720 pKa = 9.97SEE722 pKa = 4.22VLTTFDD728 pKa = 3.57FF729 pKa = 4.72
Molecular weight: 83.73 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
729 |
729 |
729 |
729.0 |
83.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.018 ± 0.0 | 1.235 ± 0.0 |
4.801 ± 0.0 | 7.27 ± 0.0 |
5.35 ± 0.0 | 5.624 ± 0.0 |
2.195 ± 0.0 | 6.584 ± 0.0 |
7.27 ± 0.0 | 10.425 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.332 ± 0.0 | 4.252 ± 0.0 |
5.761 ± 0.0 | 4.39 ± 0.0 |
4.801 ± 0.0 | 9.191 ± 0.0 |
5.35 ± 0.0 | 4.801 ± 0.0 |
1.372 ± 0.0 | 3.978 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
