
Thiobacimonas phage vB_ThpS-P1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
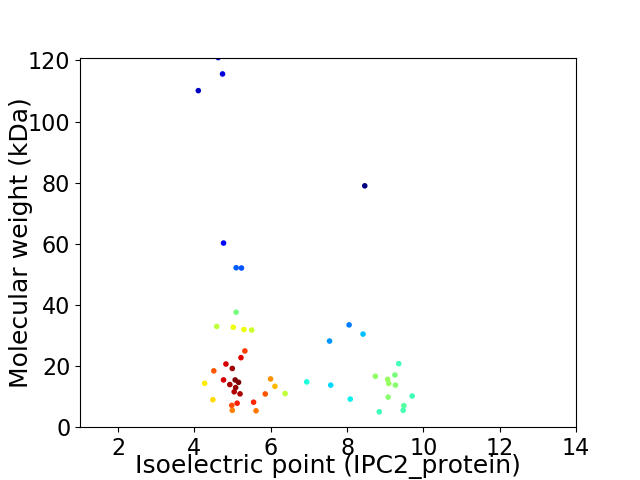
Virtual 2D-PAGE plot for 52 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B0T6D1|A0A1B0T6D1_9CAUD Tail component OS=Thiobacimonas phage vB_ThpS-P1 OX=1698523 GN=vBThpSP1_003 PE=4 SV=1
MM1 pKa = 7.95ADD3 pKa = 3.17GGVPTTEE10 pKa = 4.03LGEE13 pKa = 4.26VAGAEE18 pKa = 3.82AVLANYY24 pKa = 8.8EE25 pKa = 4.24GTTGLLSTSALASILAAAGVLAPVTVTDD53 pKa = 3.9ALGTRR58 pKa = 11.84VTALEE63 pKa = 4.16GTVIAGGSPTAGGPVAVVATGNVTLSGEE91 pKa = 4.06QTIDD95 pKa = 3.16GVTTSASRR103 pKa = 11.84VLLVGQTDD111 pKa = 3.65PAEE114 pKa = 4.06NGIWVTASGAWTRR127 pKa = 11.84ATDD130 pKa = 3.45MDD132 pKa = 4.15EE133 pKa = 4.15GAEE136 pKa = 4.09VQRR139 pKa = 11.84TYY141 pKa = 11.51VYY143 pKa = 9.68VTGGTVGRR151 pKa = 11.84GRR153 pKa = 11.84TYY155 pKa = 9.38FTGSEE160 pKa = 4.12VTTIGTDD167 pKa = 3.33DD168 pKa = 3.23IVFTEE173 pKa = 4.49FQNGNFLLTEE183 pKa = 4.72LDD185 pKa = 3.8AKK187 pKa = 11.09LDD189 pKa = 4.23DD190 pKa = 5.18ADD192 pKa = 4.63LLQSVTIAAGGTVEE206 pKa = 4.42GMANIFAAEE215 pKa = 3.68PDD217 pKa = 3.74MRR219 pKa = 11.84NGATWGDD226 pKa = 3.51VSQADD231 pKa = 4.34RR232 pKa = 11.84YY233 pKa = 11.19DD234 pKa = 3.11IGTFRR239 pKa = 11.84AAEE242 pKa = 4.16KK243 pKa = 10.51GSTGSEE249 pKa = 3.7HH250 pKa = 6.84AVYY253 pKa = 9.55TGTSATTTDD262 pKa = 2.63TWFFGAFIVSTPDD275 pKa = 3.4GVSMPSATEE284 pKa = 3.92PAPGTQTEE292 pKa = 4.12ASGTGSFSAISFASTGDD309 pKa = 2.97GGYY312 pKa = 9.89IDD314 pKa = 5.71LSDD317 pKa = 4.1DD318 pKa = 4.0ARR320 pKa = 11.84LYY322 pKa = 8.92WCYY325 pKa = 10.89GYY327 pKa = 10.28IPAGSSDD334 pKa = 4.1QICLGLASVNAGTLMSWFDD353 pKa = 4.13LRR355 pKa = 11.84LMAAEE360 pKa = 4.7PDD362 pKa = 3.68ASTVTALTQALAGAWAEE379 pKa = 4.43GVRR382 pKa = 11.84SAPLLTQASIDD393 pKa = 3.81EE394 pKa = 4.72TVLPTPITIASGGTPDD410 pKa = 5.42GYY412 pKa = 11.28DD413 pKa = 3.13NQLAGEE419 pKa = 4.19PEE421 pKa = 4.0MRR423 pKa = 11.84AEE425 pKa = 4.23ATWQTVEE432 pKa = 4.14PRR434 pKa = 11.84YY435 pKa = 9.77QSRR438 pKa = 11.84TGATRR443 pKa = 11.84AARR446 pKa = 11.84KK447 pKa = 6.9TATSSEE453 pKa = 4.22YY454 pKa = 10.85AIFSADD460 pKa = 3.22AATTTDD466 pKa = 2.49TWFFCACIVAVPPGVAMTSATVPNPGTQTDD496 pKa = 3.83GSGFAFISTSSGDD509 pKa = 3.17SGYY512 pKa = 11.08VDD514 pKa = 3.54LAEE517 pKa = 4.21GVRR520 pKa = 11.84LFWAYY525 pKa = 11.1SYY527 pKa = 10.95IPSNTTNQICVGVTGLNAGSFIGSFDD553 pKa = 3.77LRR555 pKa = 11.84LMDD558 pKa = 4.09VEE560 pKa = 4.55PSKK563 pKa = 10.8STITALTQAAAGRR576 pKa = 11.84WANGVKK582 pKa = 9.38MAEE585 pKa = 4.04VLGAQVKK592 pKa = 9.28RR593 pKa = 11.84QPTVRR598 pKa = 11.84GALSVTSAIAAVIRR612 pKa = 11.84PSKK615 pKa = 10.62LFARR619 pKa = 11.84LNPPKK624 pKa = 10.1VHH626 pKa = 6.56KK627 pKa = 10.11PSAGTIYY634 pKa = 10.5VAAPDD639 pKa = 4.09AAFLPAGSDD648 pKa = 3.29ADD650 pKa = 3.79GDD652 pKa = 4.22GSRR655 pKa = 11.84EE656 pKa = 3.89APYY659 pKa = 9.41ATLTQALSVVPSTGDD674 pKa = 3.45YY675 pKa = 11.17LILCDD680 pKa = 5.33GSFAEE685 pKa = 4.83DD686 pKa = 3.13NAGRR690 pKa = 11.84FILSGEE696 pKa = 3.82FDD698 pKa = 4.0LPVVFRR704 pKa = 11.84PYY706 pKa = 10.87VEE708 pKa = 4.82GSASITNASGTNGVINVRR726 pKa = 11.84SAAASNIQFDD736 pKa = 4.42GLTILPSTDD745 pKa = 2.64GCTTIWVNPSAGTALASNILFFNCRR770 pKa = 11.84LVQRR774 pKa = 11.84AYY776 pKa = 9.77TSAAHH781 pKa = 7.41LIRR784 pKa = 11.84FEE786 pKa = 3.77ADD788 pKa = 3.36YY789 pKa = 11.0AVKK792 pKa = 10.37GVGFVGCALVGLSGVGSTPPNIILTPANTAQVHH825 pKa = 4.66EE826 pKa = 5.35RR827 pKa = 11.84IGFWDD832 pKa = 3.94CWTEE836 pKa = 4.21GEE838 pKa = 4.31WGAFSYY844 pKa = 10.85RR845 pKa = 11.84FTGTKK850 pKa = 10.12AVTVVGNDD858 pKa = 3.05FKK860 pKa = 11.57AVVNHH865 pKa = 6.65ALLLGTDD872 pKa = 4.05SSTLSPPAISDD883 pKa = 3.58ALVIDD888 pKa = 4.21NDD890 pKa = 3.93FLAQGSNPHH899 pKa = 5.98GLEE902 pKa = 3.36IGEE905 pKa = 4.27NVSGIAALNRR915 pKa = 11.84VTSGLQGIVVKK926 pKa = 9.93GTDD929 pKa = 3.42DD930 pKa = 3.35VDD932 pKa = 4.28IIEE935 pKa = 4.3NTVRR939 pKa = 11.84LVPTAANGSALYY951 pKa = 9.61PKK953 pKa = 10.41AATNTRR959 pKa = 11.84WVGNAVIIEE968 pKa = 4.18DD969 pKa = 4.19TGLLAYY975 pKa = 9.61GFNEE979 pKa = 4.8GPDD982 pKa = 3.81GANLSGDD989 pKa = 3.47NTLSDD994 pKa = 4.07TYY996 pKa = 11.33FEE998 pKa = 4.1IHH1000 pKa = 6.21GAASQALLWSGASGSTGGATSDD1022 pKa = 3.29GNEE1025 pKa = 3.96YY1026 pKa = 11.03VLDD1029 pKa = 4.41GASLGAVRR1037 pKa = 11.84GVTVTDD1043 pKa = 3.87LAEE1046 pKa = 4.44LQAAWASDD1054 pKa = 3.39GLTGDD1059 pKa = 4.05DD1060 pKa = 4.05TANDD1064 pKa = 3.71DD1065 pKa = 4.01TSTVSAWW1072 pKa = 3.02
MM1 pKa = 7.95ADD3 pKa = 3.17GGVPTTEE10 pKa = 4.03LGEE13 pKa = 4.26VAGAEE18 pKa = 3.82AVLANYY24 pKa = 8.8EE25 pKa = 4.24GTTGLLSTSALASILAAAGVLAPVTVTDD53 pKa = 3.9ALGTRR58 pKa = 11.84VTALEE63 pKa = 4.16GTVIAGGSPTAGGPVAVVATGNVTLSGEE91 pKa = 4.06QTIDD95 pKa = 3.16GVTTSASRR103 pKa = 11.84VLLVGQTDD111 pKa = 3.65PAEE114 pKa = 4.06NGIWVTASGAWTRR127 pKa = 11.84ATDD130 pKa = 3.45MDD132 pKa = 4.15EE133 pKa = 4.15GAEE136 pKa = 4.09VQRR139 pKa = 11.84TYY141 pKa = 11.51VYY143 pKa = 9.68VTGGTVGRR151 pKa = 11.84GRR153 pKa = 11.84TYY155 pKa = 9.38FTGSEE160 pKa = 4.12VTTIGTDD167 pKa = 3.33DD168 pKa = 3.23IVFTEE173 pKa = 4.49FQNGNFLLTEE183 pKa = 4.72LDD185 pKa = 3.8AKK187 pKa = 11.09LDD189 pKa = 4.23DD190 pKa = 5.18ADD192 pKa = 4.63LLQSVTIAAGGTVEE206 pKa = 4.42GMANIFAAEE215 pKa = 3.68PDD217 pKa = 3.74MRR219 pKa = 11.84NGATWGDD226 pKa = 3.51VSQADD231 pKa = 4.34RR232 pKa = 11.84YY233 pKa = 11.19DD234 pKa = 3.11IGTFRR239 pKa = 11.84AAEE242 pKa = 4.16KK243 pKa = 10.51GSTGSEE249 pKa = 3.7HH250 pKa = 6.84AVYY253 pKa = 9.55TGTSATTTDD262 pKa = 2.63TWFFGAFIVSTPDD275 pKa = 3.4GVSMPSATEE284 pKa = 3.92PAPGTQTEE292 pKa = 4.12ASGTGSFSAISFASTGDD309 pKa = 2.97GGYY312 pKa = 9.89IDD314 pKa = 5.71LSDD317 pKa = 4.1DD318 pKa = 4.0ARR320 pKa = 11.84LYY322 pKa = 8.92WCYY325 pKa = 10.89GYY327 pKa = 10.28IPAGSSDD334 pKa = 4.1QICLGLASVNAGTLMSWFDD353 pKa = 4.13LRR355 pKa = 11.84LMAAEE360 pKa = 4.7PDD362 pKa = 3.68ASTVTALTQALAGAWAEE379 pKa = 4.43GVRR382 pKa = 11.84SAPLLTQASIDD393 pKa = 3.81EE394 pKa = 4.72TVLPTPITIASGGTPDD410 pKa = 5.42GYY412 pKa = 11.28DD413 pKa = 3.13NQLAGEE419 pKa = 4.19PEE421 pKa = 4.0MRR423 pKa = 11.84AEE425 pKa = 4.23ATWQTVEE432 pKa = 4.14PRR434 pKa = 11.84YY435 pKa = 9.77QSRR438 pKa = 11.84TGATRR443 pKa = 11.84AARR446 pKa = 11.84KK447 pKa = 6.9TATSSEE453 pKa = 4.22YY454 pKa = 10.85AIFSADD460 pKa = 3.22AATTTDD466 pKa = 2.49TWFFCACIVAVPPGVAMTSATVPNPGTQTDD496 pKa = 3.83GSGFAFISTSSGDD509 pKa = 3.17SGYY512 pKa = 11.08VDD514 pKa = 3.54LAEE517 pKa = 4.21GVRR520 pKa = 11.84LFWAYY525 pKa = 11.1SYY527 pKa = 10.95IPSNTTNQICVGVTGLNAGSFIGSFDD553 pKa = 3.77LRR555 pKa = 11.84LMDD558 pKa = 4.09VEE560 pKa = 4.55PSKK563 pKa = 10.8STITALTQAAAGRR576 pKa = 11.84WANGVKK582 pKa = 9.38MAEE585 pKa = 4.04VLGAQVKK592 pKa = 9.28RR593 pKa = 11.84QPTVRR598 pKa = 11.84GALSVTSAIAAVIRR612 pKa = 11.84PSKK615 pKa = 10.62LFARR619 pKa = 11.84LNPPKK624 pKa = 10.1VHH626 pKa = 6.56KK627 pKa = 10.11PSAGTIYY634 pKa = 10.5VAAPDD639 pKa = 4.09AAFLPAGSDD648 pKa = 3.29ADD650 pKa = 3.79GDD652 pKa = 4.22GSRR655 pKa = 11.84EE656 pKa = 3.89APYY659 pKa = 9.41ATLTQALSVVPSTGDD674 pKa = 3.45YY675 pKa = 11.17LILCDD680 pKa = 5.33GSFAEE685 pKa = 4.83DD686 pKa = 3.13NAGRR690 pKa = 11.84FILSGEE696 pKa = 3.82FDD698 pKa = 4.0LPVVFRR704 pKa = 11.84PYY706 pKa = 10.87VEE708 pKa = 4.82GSASITNASGTNGVINVRR726 pKa = 11.84SAAASNIQFDD736 pKa = 4.42GLTILPSTDD745 pKa = 2.64GCTTIWVNPSAGTALASNILFFNCRR770 pKa = 11.84LVQRR774 pKa = 11.84AYY776 pKa = 9.77TSAAHH781 pKa = 7.41LIRR784 pKa = 11.84FEE786 pKa = 3.77ADD788 pKa = 3.36YY789 pKa = 11.0AVKK792 pKa = 10.37GVGFVGCALVGLSGVGSTPPNIILTPANTAQVHH825 pKa = 4.66EE826 pKa = 5.35RR827 pKa = 11.84IGFWDD832 pKa = 3.94CWTEE836 pKa = 4.21GEE838 pKa = 4.31WGAFSYY844 pKa = 10.85RR845 pKa = 11.84FTGTKK850 pKa = 10.12AVTVVGNDD858 pKa = 3.05FKK860 pKa = 11.57AVVNHH865 pKa = 6.65ALLLGTDD872 pKa = 4.05SSTLSPPAISDD883 pKa = 3.58ALVIDD888 pKa = 4.21NDD890 pKa = 3.93FLAQGSNPHH899 pKa = 5.98GLEE902 pKa = 3.36IGEE905 pKa = 4.27NVSGIAALNRR915 pKa = 11.84VTSGLQGIVVKK926 pKa = 9.93GTDD929 pKa = 3.42DD930 pKa = 3.35VDD932 pKa = 4.28IIEE935 pKa = 4.3NTVRR939 pKa = 11.84LVPTAANGSALYY951 pKa = 9.61PKK953 pKa = 10.41AATNTRR959 pKa = 11.84WVGNAVIIEE968 pKa = 4.18DD969 pKa = 4.19TGLLAYY975 pKa = 9.61GFNEE979 pKa = 4.8GPDD982 pKa = 3.81GANLSGDD989 pKa = 3.47NTLSDD994 pKa = 4.07TYY996 pKa = 11.33FEE998 pKa = 4.1IHH1000 pKa = 6.21GAASQALLWSGASGSTGGATSDD1022 pKa = 3.29GNEE1025 pKa = 3.96YY1026 pKa = 11.03VLDD1029 pKa = 4.41GASLGAVRR1037 pKa = 11.84GVTVTDD1043 pKa = 3.87LAEE1046 pKa = 4.44LQAAWASDD1054 pKa = 3.39GLTGDD1059 pKa = 4.05DD1060 pKa = 4.05TANDD1064 pKa = 3.71DD1065 pKa = 4.01TSTVSAWW1072 pKa = 3.02
Molecular weight: 110.13 kDa
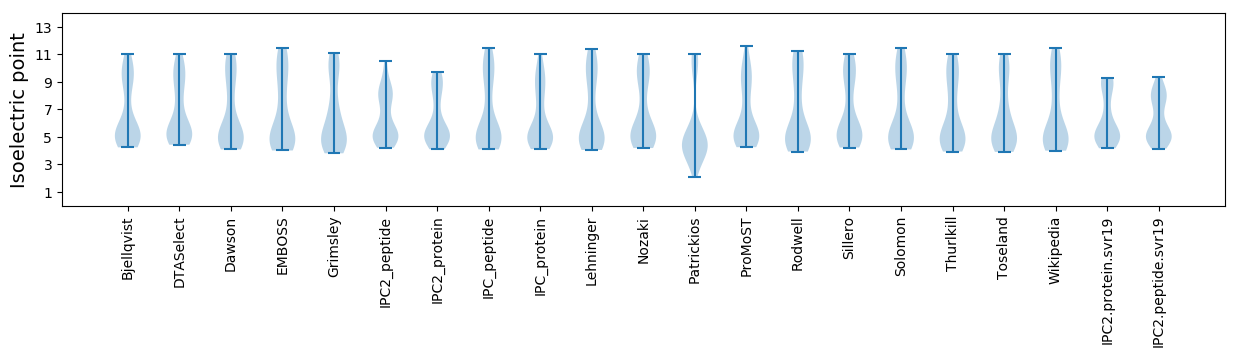
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B0T6H5|A0A1B0T6H5_9CAUD Cro/CI family transcriptional regulator OS=Thiobacimonas phage vB_ThpS-P1 OX=1698523 GN=vBThpSP1_048 PE=4 SV=1
MM1 pKa = 7.28IHH3 pKa = 6.33ATPHH7 pKa = 5.41AVRR10 pKa = 11.84RR11 pKa = 11.84PGPLICPADD20 pKa = 3.8VKK22 pKa = 10.93HH23 pKa = 5.94YY24 pKa = 9.88AAQVSRR30 pKa = 11.84GLPAEE35 pKa = 3.77QRR37 pKa = 11.84AAVAVEE43 pKa = 4.43LIRR46 pKa = 11.84SISEE50 pKa = 3.97PEE52 pKa = 3.91SFLVVRR58 pKa = 11.84DD59 pKa = 3.38AARR62 pKa = 11.84IVQGHH67 pKa = 5.92GNALLSDD74 pKa = 3.95AMRR77 pKa = 11.84VVEE80 pKa = 4.54GQTEE84 pKa = 4.34HH85 pKa = 5.44TTVASLTDD93 pKa = 3.27QSATVSPVARR103 pKa = 11.84GTAGTPRR110 pKa = 11.84SRR112 pKa = 11.84RR113 pKa = 11.84LSPLSVAWARR123 pKa = 11.84FRR125 pKa = 11.84AAEE128 pKa = 4.1RR129 pKa = 11.84RR130 pKa = 11.84FSDD133 pKa = 5.1SIWGDD138 pKa = 3.06ALAGVFALGALFGLSIILWGLQQ160 pKa = 3.05
MM1 pKa = 7.28IHH3 pKa = 6.33ATPHH7 pKa = 5.41AVRR10 pKa = 11.84RR11 pKa = 11.84PGPLICPADD20 pKa = 3.8VKK22 pKa = 10.93HH23 pKa = 5.94YY24 pKa = 9.88AAQVSRR30 pKa = 11.84GLPAEE35 pKa = 3.77QRR37 pKa = 11.84AAVAVEE43 pKa = 4.43LIRR46 pKa = 11.84SISEE50 pKa = 3.97PEE52 pKa = 3.91SFLVVRR58 pKa = 11.84DD59 pKa = 3.38AARR62 pKa = 11.84IVQGHH67 pKa = 5.92GNALLSDD74 pKa = 3.95AMRR77 pKa = 11.84VVEE80 pKa = 4.54GQTEE84 pKa = 4.34HH85 pKa = 5.44TTVASLTDD93 pKa = 3.27QSATVSPVARR103 pKa = 11.84GTAGTPRR110 pKa = 11.84SRR112 pKa = 11.84RR113 pKa = 11.84LSPLSVAWARR123 pKa = 11.84FRR125 pKa = 11.84AAEE128 pKa = 4.1RR129 pKa = 11.84RR130 pKa = 11.84FSDD133 pKa = 5.1SIWGDD138 pKa = 3.06ALAGVFALGALFGLSIILWGLQQ160 pKa = 3.05
Molecular weight: 17.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12234 |
46 |
1116 |
235.3 |
25.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.563 ± 0.425 | 0.785 ± 0.12 |
6.22 ± 0.245 | 7.021 ± 0.318 |
3.18 ± 0.161 | 8.452 ± 0.413 |
1.749 ± 0.2 | 4.733 ± 0.164 |
3.18 ± 0.313 | 8.738 ± 0.282 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.297 ± 0.209 | 2.379 ± 0.203 |
4.921 ± 0.232 | 3.245 ± 0.261 |
7.716 ± 0.461 | 5.91 ± 0.333 |
6.498 ± 0.531 | 6.523 ± 0.317 |
1.831 ± 0.118 | 2.06 ± 0.13 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
