
Streptococcus satellite phage Javan259
Taxonomy: Viruses; unclassified bacterial viruses
Average proteome isoelectric point is 5.92
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 22 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4D5ZM15|A0A4D5ZM15_9VIRU Uncharacterized protein OS=Streptococcus satellite phage Javan259 OX=2558595 GN=JavanS259_0008 PE=4 SV=1
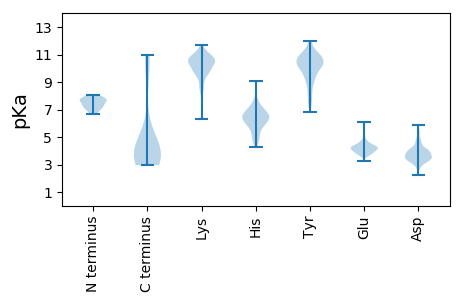
MM1 pKa = 7.69KK2 pKa = 10.26IKK4 pKa = 10.31IFYY7 pKa = 9.82QRR9 pKa = 11.84GSLTEE14 pKa = 4.12FEE16 pKa = 4.61EE17 pKa = 4.81EE18 pKa = 4.3INNFMATANVVDD30 pKa = 4.14VKK32 pKa = 10.43LTEE35 pKa = 4.13GTYY38 pKa = 11.19GDD40 pKa = 4.74YY41 pKa = 10.01EE42 pKa = 4.6TINCSLTAVIVYY54 pKa = 9.99RR55 pKa = 11.84EE56 pKa = 4.04INGVFNPVPTYY67 pKa = 11.07HH68 pKa = 6.45NN69 pKa = 3.66
MM1 pKa = 7.69KK2 pKa = 10.26IKK4 pKa = 10.31IFYY7 pKa = 9.82QRR9 pKa = 11.84GSLTEE14 pKa = 4.12FEE16 pKa = 4.61EE17 pKa = 4.81EE18 pKa = 4.3INNFMATANVVDD30 pKa = 4.14VKK32 pKa = 10.43LTEE35 pKa = 4.13GTYY38 pKa = 11.19GDD40 pKa = 4.74YY41 pKa = 10.01EE42 pKa = 4.6TINCSLTAVIVYY54 pKa = 9.99RR55 pKa = 11.84EE56 pKa = 4.04INGVFNPVPTYY67 pKa = 11.07HH68 pKa = 6.45NN69 pKa = 3.66
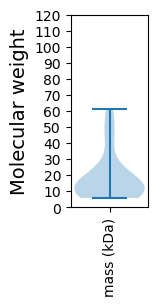
Molecular weight: 7.91 kDa
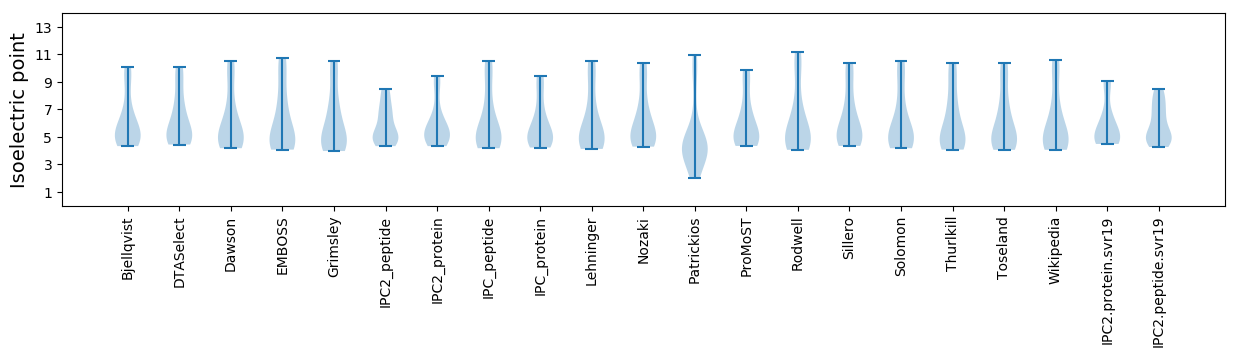
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D5ZH27|A0A4D5ZH27_9VIRU Uncharacterized protein OS=Streptococcus satellite phage Javan259 OX=2558595 GN=JavanS259_0011 PE=4 SV=1
MM1 pKa = 7.8TITEE5 pKa = 4.2YY6 pKa = 10.79KK7 pKa = 9.72KK8 pKa = 10.89KK9 pKa = 10.51NGEE12 pKa = 3.66LVYY15 pKa = 10.62RR16 pKa = 11.84SNIYY20 pKa = 10.54LGIDD24 pKa = 2.8RR25 pKa = 11.84VTGKK29 pKa = 10.3KK30 pKa = 10.03VRR32 pKa = 11.84TTITAPTKK40 pKa = 10.63KK41 pKa = 10.26LLKK44 pKa = 9.43TKK46 pKa = 10.2ARR48 pKa = 11.84QAIADD53 pKa = 4.17FEE55 pKa = 5.68ANGHH59 pKa = 5.08TIKK62 pKa = 10.68PKK64 pKa = 10.69LVVKK68 pKa = 9.48TYY70 pKa = 11.48GEE72 pKa = 4.32LAQLWWDD79 pKa = 3.92SYY81 pKa = 11.07KK82 pKa = 9.87HH83 pKa = 4.6TVKK86 pKa = 10.72PNTSCMMEE94 pKa = 4.09SFVRR98 pKa = 11.84NHH100 pKa = 6.37LLSVFADD107 pKa = 3.8YY108 pKa = 10.8PLDD111 pKa = 3.91KK112 pKa = 9.04MTTPMIQMQVNEE124 pKa = 4.36WASKK128 pKa = 9.4ANKK131 pKa = 9.35GATGAFQRR139 pKa = 11.84YY140 pKa = 8.73GLLCSLNRR148 pKa = 11.84RR149 pKa = 11.84ILQYY153 pKa = 11.17GVTLEE158 pKa = 4.36LLSFNPAKK166 pKa = 10.85NIVLPRR172 pKa = 11.84KK173 pKa = 7.84TRR175 pKa = 11.84VKK177 pKa = 10.63AKK179 pKa = 10.73DD180 pKa = 3.58KK181 pKa = 10.73IKK183 pKa = 10.96HH184 pKa = 6.26LDD186 pKa = 3.4QDD188 pKa = 3.76NLKK191 pKa = 10.99AFLTYY196 pKa = 10.62LSRR199 pKa = 11.84LEE201 pKa = 3.78PTYY204 pKa = 11.11QHH206 pKa = 6.23MFEE209 pKa = 3.89IMLYY213 pKa = 8.55KK214 pKa = 10.18TLLATGCRR222 pKa = 11.84ISEE225 pKa = 4.2LLALEE230 pKa = 4.26WSDD233 pKa = 3.7IDD235 pKa = 4.13FDD237 pKa = 4.03EE238 pKa = 4.74AQISITKK245 pKa = 8.33TLNRR249 pKa = 11.84YY250 pKa = 7.38RR251 pKa = 11.84TVNSPKK257 pKa = 9.29SKK259 pKa = 10.5SSIRR263 pKa = 11.84VIDD266 pKa = 3.52IDD268 pKa = 3.91QATLKK273 pKa = 9.54MLKK276 pKa = 9.83SYY278 pKa = 10.34RR279 pKa = 11.84QQQRR283 pKa = 11.84LEE285 pKa = 4.05AGYY288 pKa = 9.45QPKK291 pKa = 10.07IVFSPITDD299 pKa = 4.04TYY301 pKa = 11.69AKK303 pKa = 10.12VASLRR308 pKa = 11.84QRR310 pKa = 11.84LEE312 pKa = 3.48RR313 pKa = 11.84HH314 pKa = 5.19FRR316 pKa = 11.84LAGVEE321 pKa = 3.86NVGFHH326 pKa = 6.51GFRR329 pKa = 11.84HH330 pKa = 4.86THH332 pKa = 6.74ASLLLNAGIPYY343 pKa = 10.01KK344 pKa = 10.28EE345 pKa = 3.98LQHH348 pKa = 7.05RR349 pKa = 11.84LGHH352 pKa = 5.92NNISTTMDD360 pKa = 2.78IYY362 pKa = 11.63GHH364 pKa = 6.4LSEE367 pKa = 4.86MNKK370 pKa = 9.94KK371 pKa = 9.68SAVSFFEE378 pKa = 3.98NAINEE383 pKa = 4.37LKK385 pKa = 10.82VV386 pKa = 3.17
MM1 pKa = 7.8TITEE5 pKa = 4.2YY6 pKa = 10.79KK7 pKa = 9.72KK8 pKa = 10.89KK9 pKa = 10.51NGEE12 pKa = 3.66LVYY15 pKa = 10.62RR16 pKa = 11.84SNIYY20 pKa = 10.54LGIDD24 pKa = 2.8RR25 pKa = 11.84VTGKK29 pKa = 10.3KK30 pKa = 10.03VRR32 pKa = 11.84TTITAPTKK40 pKa = 10.63KK41 pKa = 10.26LLKK44 pKa = 9.43TKK46 pKa = 10.2ARR48 pKa = 11.84QAIADD53 pKa = 4.17FEE55 pKa = 5.68ANGHH59 pKa = 5.08TIKK62 pKa = 10.68PKK64 pKa = 10.69LVVKK68 pKa = 9.48TYY70 pKa = 11.48GEE72 pKa = 4.32LAQLWWDD79 pKa = 3.92SYY81 pKa = 11.07KK82 pKa = 9.87HH83 pKa = 4.6TVKK86 pKa = 10.72PNTSCMMEE94 pKa = 4.09SFVRR98 pKa = 11.84NHH100 pKa = 6.37LLSVFADD107 pKa = 3.8YY108 pKa = 10.8PLDD111 pKa = 3.91KK112 pKa = 9.04MTTPMIQMQVNEE124 pKa = 4.36WASKK128 pKa = 9.4ANKK131 pKa = 9.35GATGAFQRR139 pKa = 11.84YY140 pKa = 8.73GLLCSLNRR148 pKa = 11.84RR149 pKa = 11.84ILQYY153 pKa = 11.17GVTLEE158 pKa = 4.36LLSFNPAKK166 pKa = 10.85NIVLPRR172 pKa = 11.84KK173 pKa = 7.84TRR175 pKa = 11.84VKK177 pKa = 10.63AKK179 pKa = 10.73DD180 pKa = 3.58KK181 pKa = 10.73IKK183 pKa = 10.96HH184 pKa = 6.26LDD186 pKa = 3.4QDD188 pKa = 3.76NLKK191 pKa = 10.99AFLTYY196 pKa = 10.62LSRR199 pKa = 11.84LEE201 pKa = 3.78PTYY204 pKa = 11.11QHH206 pKa = 6.23MFEE209 pKa = 3.89IMLYY213 pKa = 8.55KK214 pKa = 10.18TLLATGCRR222 pKa = 11.84ISEE225 pKa = 4.2LLALEE230 pKa = 4.26WSDD233 pKa = 3.7IDD235 pKa = 4.13FDD237 pKa = 4.03EE238 pKa = 4.74AQISITKK245 pKa = 8.33TLNRR249 pKa = 11.84YY250 pKa = 7.38RR251 pKa = 11.84TVNSPKK257 pKa = 9.29SKK259 pKa = 10.5SSIRR263 pKa = 11.84VIDD266 pKa = 3.52IDD268 pKa = 3.91QATLKK273 pKa = 9.54MLKK276 pKa = 9.83SYY278 pKa = 10.34RR279 pKa = 11.84QQQRR283 pKa = 11.84LEE285 pKa = 4.05AGYY288 pKa = 9.45QPKK291 pKa = 10.07IVFSPITDD299 pKa = 4.04TYY301 pKa = 11.69AKK303 pKa = 10.12VASLRR308 pKa = 11.84QRR310 pKa = 11.84LEE312 pKa = 3.48RR313 pKa = 11.84HH314 pKa = 5.19FRR316 pKa = 11.84LAGVEE321 pKa = 3.86NVGFHH326 pKa = 6.51GFRR329 pKa = 11.84HH330 pKa = 4.86THH332 pKa = 6.74ASLLLNAGIPYY343 pKa = 10.01KK344 pKa = 10.28EE345 pKa = 3.98LQHH348 pKa = 7.05RR349 pKa = 11.84LGHH352 pKa = 5.92NNISTTMDD360 pKa = 2.78IYY362 pKa = 11.63GHH364 pKa = 6.4LSEE367 pKa = 4.86MNKK370 pKa = 9.94KK371 pKa = 9.68SAVSFFEE378 pKa = 3.98NAINEE383 pKa = 4.37LKK385 pKa = 10.82VV386 pKa = 3.17
Molecular weight: 44.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3482 |
49 |
535 |
158.3 |
18.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.83 ± 0.326 | 0.574 ± 0.173 |
6.318 ± 0.397 | 8.702 ± 0.727 |
4.423 ± 0.508 | 4.968 ± 0.408 |
1.895 ± 0.261 | 6.72 ± 0.309 |
8.845 ± 0.467 | 9.075 ± 0.508 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.556 ± 0.239 | 5.227 ± 0.338 |
2.298 ± 0.201 | 4.193 ± 0.248 |
4.94 ± 0.46 | 5.313 ± 0.39 |
6.519 ± 0.391 | 5.773 ± 0.34 |
1.149 ± 0.228 | 4.681 ± 0.236 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
