
Rhodopirellula sp. MGV
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Planctomycetia; Pirellulales; Pirellulaceae; Rhodopirellula; unclassified Rhodopirellula
Average proteome isoelectric point is 6.05
Get precalculated fractions of proteins
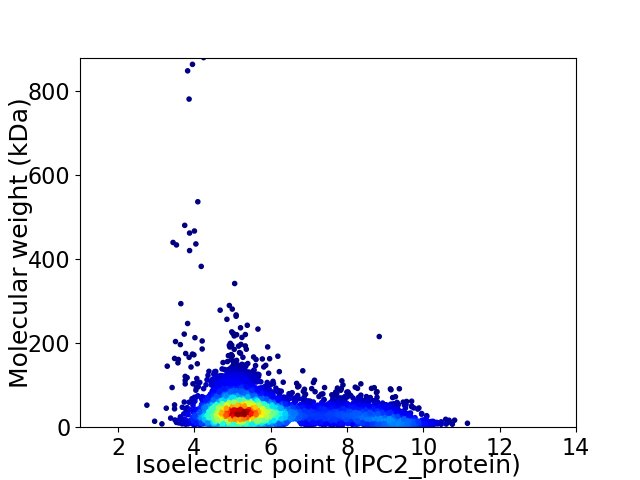
Virtual 2D-PAGE plot for 5273 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
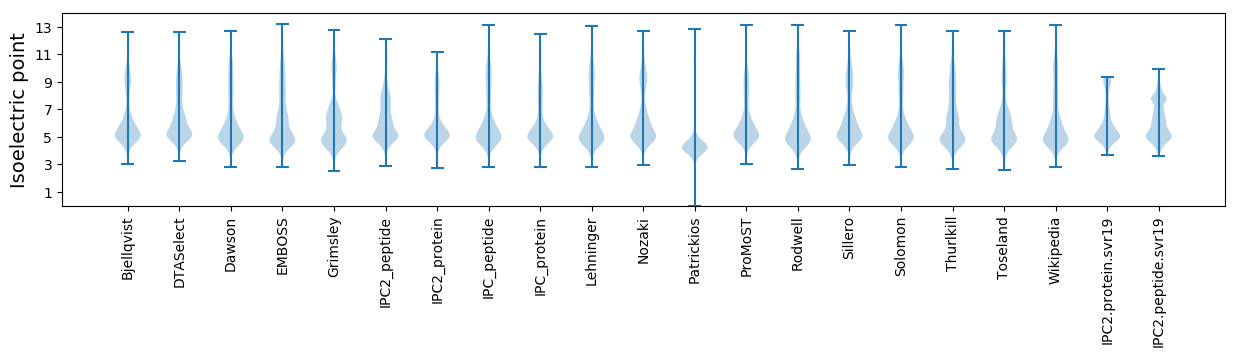
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A255QV90|A0A255QV90_9BACT Phosphate import ATP-binding protein PstB OS=Rhodopirellula sp. MGV OX=2023130 GN=pstB PE=3 SV=1
MM1 pKa = 7.27ARR3 pKa = 11.84KK4 pKa = 9.77LSFEE8 pKa = 3.97SMEE11 pKa = 3.98ARR13 pKa = 11.84RR14 pKa = 11.84LLAADD19 pKa = 3.2VAFNHH24 pKa = 6.29EE25 pKa = 4.35EE26 pKa = 3.93PTFEE30 pKa = 4.12VASDD34 pKa = 3.42ISAKK38 pKa = 10.57AFADD42 pKa = 3.09IGNLDD47 pKa = 3.42GTAIISSSLGWFNSYY62 pKa = 11.02DD63 pKa = 3.65RR64 pKa = 11.84IGFSVEE70 pKa = 3.54RR71 pKa = 11.84EE72 pKa = 3.6ADD74 pKa = 3.18VSIRR78 pKa = 11.84LNSFWTDD85 pKa = 2.26VDD87 pKa = 5.26LYY89 pKa = 10.23LTDD92 pKa = 3.06SRR94 pKa = 11.84GYY96 pKa = 10.53VIDD99 pKa = 4.43YY100 pKa = 9.57SVNWGTTAEE109 pKa = 4.58SIDD112 pKa = 3.25ITVNPGNYY120 pKa = 9.14YY121 pKa = 9.49VWSLATSWYY130 pKa = 5.03TTGYY134 pKa = 10.58RR135 pKa = 11.84MEE137 pKa = 4.26LTADD141 pKa = 3.56QVPEE145 pKa = 4.19PLPEE149 pKa = 4.29PEE151 pKa = 4.3PTPDD155 pKa = 3.37AGVPADD161 pKa = 4.3GVSPLPDD168 pKa = 2.84VAYY171 pKa = 10.59FGGSNEE177 pKa = 3.83WGLNAVGAPEE187 pKa = 3.93AWAAGYY193 pKa = 7.83TGQGVTVAVIDD204 pKa = 3.7TGVDD208 pKa = 3.35LDD210 pKa = 5.13HH211 pKa = 7.73PDD213 pKa = 3.85LYY215 pKa = 11.7SNIYY219 pKa = 9.37VNPGEE224 pKa = 4.11IAGNGIDD231 pKa = 5.14DD232 pKa = 5.04DD233 pKa = 5.1GNGYY237 pKa = 10.18VDD239 pKa = 4.78DD240 pKa = 3.73VHH242 pKa = 7.97GFDD245 pKa = 5.68FADD248 pKa = 5.07FDD250 pKa = 4.93ADD252 pKa = 3.79PNDD255 pKa = 3.57VHH257 pKa = 7.37GHH259 pKa = 4.6GTHH262 pKa = 5.72VAGTIAALDD271 pKa = 4.05NGWGATGVAPEE282 pKa = 4.03ATILPVKK289 pKa = 10.63VLGDD293 pKa = 3.67NGSGSSTSVAAGIRR307 pKa = 11.84YY308 pKa = 8.6AADD311 pKa = 3.07MGADD315 pKa = 4.15IINLSLGGGYY325 pKa = 10.3SSDD328 pKa = 3.2IEE330 pKa = 4.32SAINYY335 pKa = 8.38ARR337 pKa = 11.84TLGSLVIAAAGNEE350 pKa = 4.25YY351 pKa = 10.95ASMPGFPARR360 pKa = 11.84FSASYY365 pKa = 11.42DD366 pKa = 3.36NVLSVGAYY374 pKa = 10.28DD375 pKa = 3.49SGSNIASFSNDD386 pKa = 2.88VGGSGAVQVDD396 pKa = 3.78APGVGIFSTYY406 pKa = 10.22VGGGFGTMSGTSMATPHH423 pKa = 5.56VVGVAALTLSANPSLSPAALRR444 pKa = 11.84DD445 pKa = 3.75LLVSGVNGHH454 pKa = 7.11ANGSDD459 pKa = 3.5GLGKK463 pKa = 10.51ASTIYY468 pKa = 10.52SVAYY472 pKa = 9.79AAAGVSGSSTSGSGSTSTSGGSGSVGAASVGDD504 pKa = 3.87AGGSTLAGLSDD515 pKa = 4.87SIEE518 pKa = 4.49DD519 pKa = 3.9DD520 pKa = 3.11ATNADD525 pKa = 3.97RR526 pKa = 11.84VFEE529 pKa = 4.3SDD531 pKa = 3.6VQFDD535 pKa = 3.59SSPRR539 pKa = 11.84LARR542 pKa = 11.84SNRR545 pKa = 11.84PSVAPQPEE553 pKa = 4.28DD554 pKa = 3.1TDD556 pKa = 3.97AALSDD561 pKa = 3.83FFGDD565 pKa = 3.38SDD567 pKa = 4.03SDD569 pKa = 3.98NEE571 pKa = 4.22FGSDD575 pKa = 3.37RR576 pKa = 11.84EE577 pKa = 4.02VDD579 pKa = 3.57LGLLVLAA586 pKa = 5.61
MM1 pKa = 7.27ARR3 pKa = 11.84KK4 pKa = 9.77LSFEE8 pKa = 3.97SMEE11 pKa = 3.98ARR13 pKa = 11.84RR14 pKa = 11.84LLAADD19 pKa = 3.2VAFNHH24 pKa = 6.29EE25 pKa = 4.35EE26 pKa = 3.93PTFEE30 pKa = 4.12VASDD34 pKa = 3.42ISAKK38 pKa = 10.57AFADD42 pKa = 3.09IGNLDD47 pKa = 3.42GTAIISSSLGWFNSYY62 pKa = 11.02DD63 pKa = 3.65RR64 pKa = 11.84IGFSVEE70 pKa = 3.54RR71 pKa = 11.84EE72 pKa = 3.6ADD74 pKa = 3.18VSIRR78 pKa = 11.84LNSFWTDD85 pKa = 2.26VDD87 pKa = 5.26LYY89 pKa = 10.23LTDD92 pKa = 3.06SRR94 pKa = 11.84GYY96 pKa = 10.53VIDD99 pKa = 4.43YY100 pKa = 9.57SVNWGTTAEE109 pKa = 4.58SIDD112 pKa = 3.25ITVNPGNYY120 pKa = 9.14YY121 pKa = 9.49VWSLATSWYY130 pKa = 5.03TTGYY134 pKa = 10.58RR135 pKa = 11.84MEE137 pKa = 4.26LTADD141 pKa = 3.56QVPEE145 pKa = 4.19PLPEE149 pKa = 4.29PEE151 pKa = 4.3PTPDD155 pKa = 3.37AGVPADD161 pKa = 4.3GVSPLPDD168 pKa = 2.84VAYY171 pKa = 10.59FGGSNEE177 pKa = 3.83WGLNAVGAPEE187 pKa = 3.93AWAAGYY193 pKa = 7.83TGQGVTVAVIDD204 pKa = 3.7TGVDD208 pKa = 3.35LDD210 pKa = 5.13HH211 pKa = 7.73PDD213 pKa = 3.85LYY215 pKa = 11.7SNIYY219 pKa = 9.37VNPGEE224 pKa = 4.11IAGNGIDD231 pKa = 5.14DD232 pKa = 5.04DD233 pKa = 5.1GNGYY237 pKa = 10.18VDD239 pKa = 4.78DD240 pKa = 3.73VHH242 pKa = 7.97GFDD245 pKa = 5.68FADD248 pKa = 5.07FDD250 pKa = 4.93ADD252 pKa = 3.79PNDD255 pKa = 3.57VHH257 pKa = 7.37GHH259 pKa = 4.6GTHH262 pKa = 5.72VAGTIAALDD271 pKa = 4.05NGWGATGVAPEE282 pKa = 4.03ATILPVKK289 pKa = 10.63VLGDD293 pKa = 3.67NGSGSSTSVAAGIRR307 pKa = 11.84YY308 pKa = 8.6AADD311 pKa = 3.07MGADD315 pKa = 4.15IINLSLGGGYY325 pKa = 10.3SSDD328 pKa = 3.2IEE330 pKa = 4.32SAINYY335 pKa = 8.38ARR337 pKa = 11.84TLGSLVIAAAGNEE350 pKa = 4.25YY351 pKa = 10.95ASMPGFPARR360 pKa = 11.84FSASYY365 pKa = 11.42DD366 pKa = 3.36NVLSVGAYY374 pKa = 10.28DD375 pKa = 3.49SGSNIASFSNDD386 pKa = 2.88VGGSGAVQVDD396 pKa = 3.78APGVGIFSTYY406 pKa = 10.22VGGGFGTMSGTSMATPHH423 pKa = 5.56VVGVAALTLSANPSLSPAALRR444 pKa = 11.84DD445 pKa = 3.75LLVSGVNGHH454 pKa = 7.11ANGSDD459 pKa = 3.5GLGKK463 pKa = 10.51ASTIYY468 pKa = 10.52SVAYY472 pKa = 9.79AAAGVSGSSTSGSGSTSTSGGSGSVGAASVGDD504 pKa = 3.87AGGSTLAGLSDD515 pKa = 4.87SIEE518 pKa = 4.49DD519 pKa = 3.9DD520 pKa = 3.11ATNADD525 pKa = 3.97RR526 pKa = 11.84VFEE529 pKa = 4.3SDD531 pKa = 3.6VQFDD535 pKa = 3.59SSPRR539 pKa = 11.84LARR542 pKa = 11.84SNRR545 pKa = 11.84PSVAPQPEE553 pKa = 4.28DD554 pKa = 3.1TDD556 pKa = 3.97AALSDD561 pKa = 3.83FFGDD565 pKa = 3.38SDD567 pKa = 4.03SDD569 pKa = 3.98NEE571 pKa = 4.22FGSDD575 pKa = 3.37RR576 pKa = 11.84EE577 pKa = 4.02VDD579 pKa = 3.57LGLLVLAA586 pKa = 5.61
Molecular weight: 59.89 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A255R7Q9|A0A255R7Q9_9BACT Uncharacterized protein OS=Rhodopirellula sp. MGV OX=2023130 GN=CGZ80_07480 PE=4 SV=1
MM1 pKa = 7.17TLHH4 pKa = 7.1RR5 pKa = 11.84VPIPRR10 pKa = 11.84RR11 pKa = 11.84NTVAKK16 pKa = 10.43KK17 pKa = 10.3KK18 pKa = 8.91ATKK21 pKa = 10.26KK22 pKa = 10.4KK23 pKa = 8.66ATKK26 pKa = 10.15KK27 pKa = 10.11KK28 pKa = 9.49AAKK31 pKa = 9.89KK32 pKa = 10.03KK33 pKa = 8.36ATKK36 pKa = 10.13KK37 pKa = 9.97VATKK41 pKa = 10.49KK42 pKa = 9.02ATKK45 pKa = 10.21KK46 pKa = 10.44KK47 pKa = 8.63ATKK50 pKa = 10.25KK51 pKa = 10.39KK52 pKa = 8.55ATKK55 pKa = 10.25KK56 pKa = 10.39KK57 pKa = 8.55ATKK60 pKa = 10.25KK61 pKa = 10.39KK62 pKa = 8.55ATKK65 pKa = 10.25KK66 pKa = 10.39KK67 pKa = 8.55ATKK70 pKa = 10.25KK71 pKa = 10.39KK72 pKa = 8.66ATKK75 pKa = 10.09KK76 pKa = 9.83KK77 pKa = 9.18AAKK80 pKa = 9.59KK81 pKa = 10.04RR82 pKa = 11.84RR83 pKa = 3.58
MM1 pKa = 7.17TLHH4 pKa = 7.1RR5 pKa = 11.84VPIPRR10 pKa = 11.84RR11 pKa = 11.84NTVAKK16 pKa = 10.43KK17 pKa = 10.3KK18 pKa = 8.91ATKK21 pKa = 10.26KK22 pKa = 10.4KK23 pKa = 8.66ATKK26 pKa = 10.15KK27 pKa = 10.11KK28 pKa = 9.49AAKK31 pKa = 9.89KK32 pKa = 10.03KK33 pKa = 8.36ATKK36 pKa = 10.13KK37 pKa = 9.97VATKK41 pKa = 10.49KK42 pKa = 9.02ATKK45 pKa = 10.21KK46 pKa = 10.44KK47 pKa = 8.63ATKK50 pKa = 10.25KK51 pKa = 10.39KK52 pKa = 8.55ATKK55 pKa = 10.25KK56 pKa = 10.39KK57 pKa = 8.55ATKK60 pKa = 10.25KK61 pKa = 10.39KK62 pKa = 8.55ATKK65 pKa = 10.25KK66 pKa = 10.39KK67 pKa = 8.55ATKK70 pKa = 10.25KK71 pKa = 10.39KK72 pKa = 8.66ATKK75 pKa = 10.09KK76 pKa = 9.83KK77 pKa = 9.18AAKK80 pKa = 9.59KK81 pKa = 10.04RR82 pKa = 11.84RR83 pKa = 3.58
Molecular weight: 9.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2047646 |
30 |
8214 |
388.3 |
42.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.497 ± 0.032 | 1.142 ± 0.016 |
6.492 ± 0.035 | 6.133 ± 0.038 |
3.746 ± 0.023 | 7.484 ± 0.041 |
2.117 ± 0.021 | 5.288 ± 0.021 |
3.495 ± 0.028 | 9.593 ± 0.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.224 ± 0.017 | 3.36 ± 0.024 |
4.95 ± 0.027 | 4.239 ± 0.023 |
6.546 ± 0.04 | 7.04 ± 0.031 |
5.682 ± 0.044 | 7.201 ± 0.031 |
1.409 ± 0.015 | 2.362 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
