
Streptomyces sp. A0642
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces; unclassified Streptomyces
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 6934 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
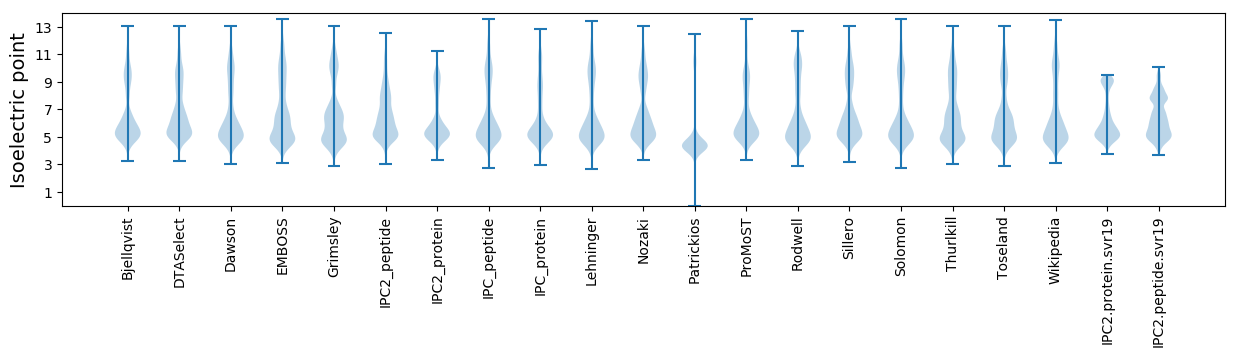
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4S2V1I5|A0A4S2V1I5_9ACTN Flavoprotein oxidoreductase OS=Streptomyces sp. A0642 OX=2563100 GN=E6R60_00210 PE=4 SV=1
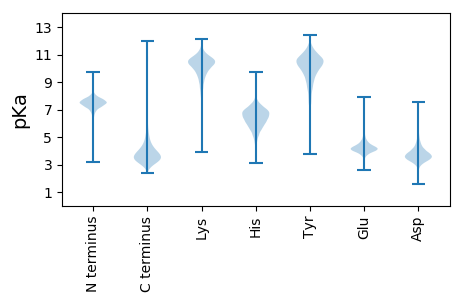
MM1 pKa = 7.51SNEE4 pKa = 3.79DD5 pKa = 3.67HH6 pKa = 7.38ADD8 pKa = 3.15QHH10 pKa = 6.35GRR12 pKa = 11.84WEE14 pKa = 4.0PTPQGGEE21 pKa = 3.8YY22 pKa = 10.25DD23 pKa = 3.75AEE25 pKa = 4.03ATAFVHH31 pKa = 6.98LPPEE35 pKa = 4.33DD36 pKa = 3.69LANIPLAAPGQGYY49 pKa = 8.52VPPMILPLTPAAGLDD64 pKa = 3.66PAATGNWVVATPDD77 pKa = 3.36QPDD80 pKa = 3.54EE81 pKa = 4.19QEE83 pKa = 4.53PGAGPAAVHH92 pKa = 6.12WPDD95 pKa = 4.07PNQQQTSYY103 pKa = 10.52GYY105 pKa = 8.53PQASQDD111 pKa = 3.59YY112 pKa = 8.69PDD114 pKa = 5.6PYY116 pKa = 10.41TGTQAATGQWNFAEE130 pKa = 4.13NEE132 pKa = 4.45GPVEE136 pKa = 4.18AAGHH140 pKa = 5.79TGQWTIPVADD150 pKa = 3.97GEE152 pKa = 4.37LPEE155 pKa = 4.33EE156 pKa = 4.39SGEE159 pKa = 4.17FSASALAAGWYY170 pKa = 8.69QDD172 pKa = 3.46RR173 pKa = 11.84TPPATLPGGAPAPWATQEE191 pKa = 4.02PQPAATEE198 pKa = 4.31TPARR202 pKa = 11.84GTDD205 pKa = 3.72LGPLPEE211 pKa = 4.73SAAEE215 pKa = 4.35APAEE219 pKa = 4.31APAEE223 pKa = 4.33APAEE227 pKa = 4.31APAEE231 pKa = 4.17EE232 pKa = 4.99PGADD236 pKa = 3.31PVADD240 pKa = 3.94GTAEE244 pKa = 4.11AVPGTPDD251 pKa = 3.61DD252 pKa = 3.95LAHH255 pKa = 7.16RR256 pKa = 11.84SPAEE260 pKa = 3.66EE261 pKa = 4.17TAPDD265 pKa = 4.12TAPEE269 pKa = 4.38DD270 pKa = 4.08APEE273 pKa = 4.02QPEE276 pKa = 3.91ASAPDD281 pKa = 3.85GVVPDD286 pKa = 5.17DD287 pKa = 4.27AASDD291 pKa = 4.38GPAPDD296 pKa = 5.07DD297 pKa = 4.1AVADD301 pKa = 4.29APPAAPAFDD310 pKa = 4.73LPSEE314 pKa = 4.62HH315 pKa = 7.04PAASYY320 pKa = 10.25VLHH323 pKa = 6.66VNGADD328 pKa = 4.49RR329 pKa = 11.84PVTDD333 pKa = 3.02AWIGEE338 pKa = 4.15SLLYY342 pKa = 10.18VLRR345 pKa = 11.84EE346 pKa = 3.94RR347 pKa = 11.84LGLAGAKK354 pKa = 9.72DD355 pKa = 3.75GCSQGEE361 pKa = 4.12CGACNVQVDD370 pKa = 3.94GRR372 pKa = 11.84LVASCLVPAATAAGSEE388 pKa = 4.08VRR390 pKa = 11.84TVEE393 pKa = 3.9GLAVDD398 pKa = 4.82GEE400 pKa = 4.31PSDD403 pKa = 3.93VQRR406 pKa = 11.84ALADD410 pKa = 3.82CGAVQCGFCIPGMAMTVHH428 pKa = 7.14DD429 pKa = 5.06LLEE432 pKa = 4.83GNHH435 pKa = 6.43APSEE439 pKa = 4.33LEE441 pKa = 3.87TRR443 pKa = 11.84QALCGNLCRR452 pKa = 11.84CSGYY456 pKa = 10.39RR457 pKa = 11.84GVLDD461 pKa = 3.98AVNEE465 pKa = 4.35VIAGRR470 pKa = 11.84EE471 pKa = 3.92AASEE475 pKa = 4.1GAAAPEE481 pKa = 4.71PGSGDD486 pKa = 3.48PDD488 pKa = 3.86GARR491 pKa = 11.84IPHH494 pKa = 5.7QAAPGAGSVQAHH506 pKa = 5.78LQDD509 pKa = 3.62GGMAA513 pKa = 3.92
MM1 pKa = 7.51SNEE4 pKa = 3.79DD5 pKa = 3.67HH6 pKa = 7.38ADD8 pKa = 3.15QHH10 pKa = 6.35GRR12 pKa = 11.84WEE14 pKa = 4.0PTPQGGEE21 pKa = 3.8YY22 pKa = 10.25DD23 pKa = 3.75AEE25 pKa = 4.03ATAFVHH31 pKa = 6.98LPPEE35 pKa = 4.33DD36 pKa = 3.69LANIPLAAPGQGYY49 pKa = 8.52VPPMILPLTPAAGLDD64 pKa = 3.66PAATGNWVVATPDD77 pKa = 3.36QPDD80 pKa = 3.54EE81 pKa = 4.19QEE83 pKa = 4.53PGAGPAAVHH92 pKa = 6.12WPDD95 pKa = 4.07PNQQQTSYY103 pKa = 10.52GYY105 pKa = 8.53PQASQDD111 pKa = 3.59YY112 pKa = 8.69PDD114 pKa = 5.6PYY116 pKa = 10.41TGTQAATGQWNFAEE130 pKa = 4.13NEE132 pKa = 4.45GPVEE136 pKa = 4.18AAGHH140 pKa = 5.79TGQWTIPVADD150 pKa = 3.97GEE152 pKa = 4.37LPEE155 pKa = 4.33EE156 pKa = 4.39SGEE159 pKa = 4.17FSASALAAGWYY170 pKa = 8.69QDD172 pKa = 3.46RR173 pKa = 11.84TPPATLPGGAPAPWATQEE191 pKa = 4.02PQPAATEE198 pKa = 4.31TPARR202 pKa = 11.84GTDD205 pKa = 3.72LGPLPEE211 pKa = 4.73SAAEE215 pKa = 4.35APAEE219 pKa = 4.31APAEE223 pKa = 4.33APAEE227 pKa = 4.31APAEE231 pKa = 4.17EE232 pKa = 4.99PGADD236 pKa = 3.31PVADD240 pKa = 3.94GTAEE244 pKa = 4.11AVPGTPDD251 pKa = 3.61DD252 pKa = 3.95LAHH255 pKa = 7.16RR256 pKa = 11.84SPAEE260 pKa = 3.66EE261 pKa = 4.17TAPDD265 pKa = 4.12TAPEE269 pKa = 4.38DD270 pKa = 4.08APEE273 pKa = 4.02QPEE276 pKa = 3.91ASAPDD281 pKa = 3.85GVVPDD286 pKa = 5.17DD287 pKa = 4.27AASDD291 pKa = 4.38GPAPDD296 pKa = 5.07DD297 pKa = 4.1AVADD301 pKa = 4.29APPAAPAFDD310 pKa = 4.73LPSEE314 pKa = 4.62HH315 pKa = 7.04PAASYY320 pKa = 10.25VLHH323 pKa = 6.66VNGADD328 pKa = 4.49RR329 pKa = 11.84PVTDD333 pKa = 3.02AWIGEE338 pKa = 4.15SLLYY342 pKa = 10.18VLRR345 pKa = 11.84EE346 pKa = 3.94RR347 pKa = 11.84LGLAGAKK354 pKa = 9.72DD355 pKa = 3.75GCSQGEE361 pKa = 4.12CGACNVQVDD370 pKa = 3.94GRR372 pKa = 11.84LVASCLVPAATAAGSEE388 pKa = 4.08VRR390 pKa = 11.84TVEE393 pKa = 3.9GLAVDD398 pKa = 4.82GEE400 pKa = 4.31PSDD403 pKa = 3.93VQRR406 pKa = 11.84ALADD410 pKa = 3.82CGAVQCGFCIPGMAMTVHH428 pKa = 7.14DD429 pKa = 5.06LLEE432 pKa = 4.83GNHH435 pKa = 6.43APSEE439 pKa = 4.33LEE441 pKa = 3.87TRR443 pKa = 11.84QALCGNLCRR452 pKa = 11.84CSGYY456 pKa = 10.39RR457 pKa = 11.84GVLDD461 pKa = 3.98AVNEE465 pKa = 4.35VIAGRR470 pKa = 11.84EE471 pKa = 3.92AASEE475 pKa = 4.1GAAAPEE481 pKa = 4.71PGSGDD486 pKa = 3.48PDD488 pKa = 3.86GARR491 pKa = 11.84IPHH494 pKa = 5.7QAAPGAGSVQAHH506 pKa = 5.78LQDD509 pKa = 3.62GGMAA513 pKa = 3.92
Molecular weight: 52.03 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4S2UVC2|A0A4S2UVC2_9ACTN Aminoglycoside phosphotransferase OS=Streptomyces sp. A0642 OX=2563100 GN=E6R60_13265 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILASRR35 pKa = 11.84RR36 pKa = 11.84GKK38 pKa = 10.5GRR40 pKa = 11.84ASLSAA45 pKa = 3.83
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILASRR35 pKa = 11.84RR36 pKa = 11.84GKK38 pKa = 10.5GRR40 pKa = 11.84ASLSAA45 pKa = 3.83
Molecular weight: 5.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2322785 |
18 |
6264 |
335.0 |
35.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.739 ± 0.047 | 0.772 ± 0.009 |
5.942 ± 0.024 | 5.624 ± 0.033 |
2.731 ± 0.017 | 9.701 ± 0.029 |
2.271 ± 0.016 | 3.203 ± 0.019 |
2.2 ± 0.027 | 10.207 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.773 ± 0.014 | 1.795 ± 0.018 |
6.088 ± 0.027 | 2.774 ± 0.019 |
7.828 ± 0.037 | 5.198 ± 0.023 |
6.167 ± 0.032 | 8.358 ± 0.028 |
1.519 ± 0.012 | 2.109 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
