
Odonata-associated circular virus-16
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 7.85
Get precalculated fractions of proteins
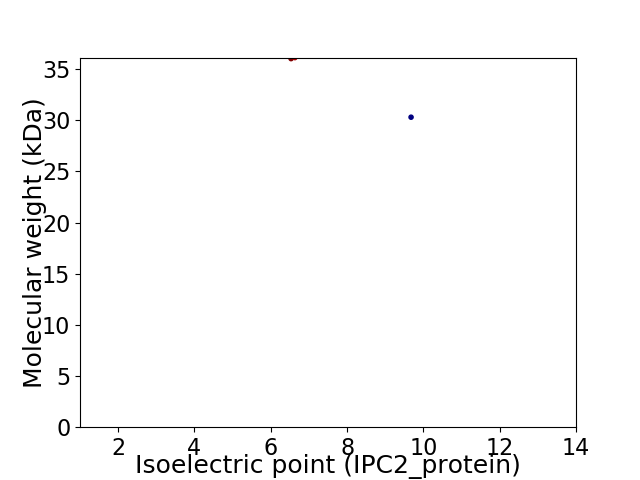
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
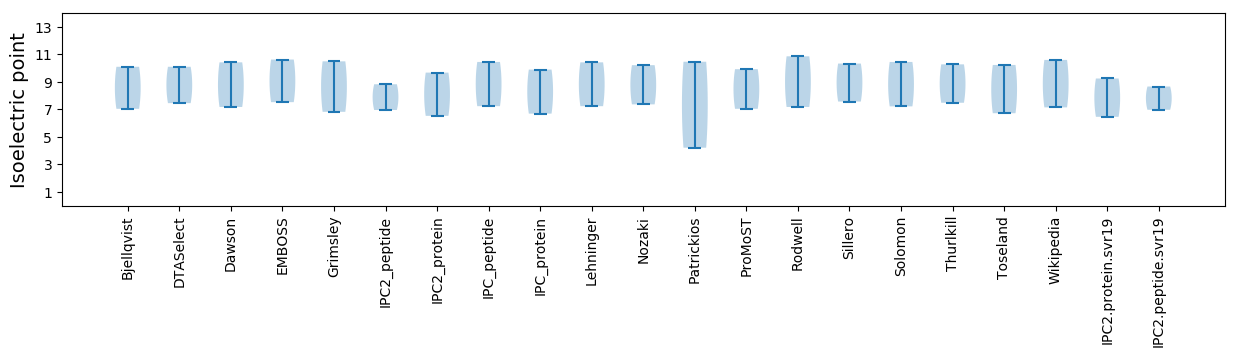
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4UGW2|A0A0B4UGW2_9VIRU Putative capsid protein OS=Odonata-associated circular virus-16 OX=1592116 PE=4 SV=1
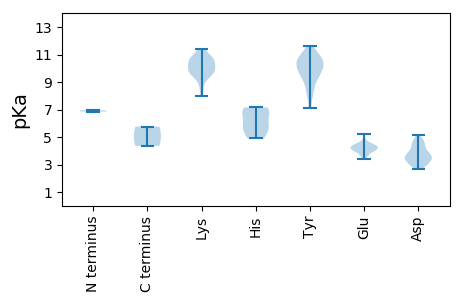
MM1 pKa = 6.95PRR3 pKa = 11.84KK4 pKa = 9.13MEE6 pKa = 4.27SLEE9 pKa = 4.04SSGGGNSIPPPIKK22 pKa = 9.83EE23 pKa = 3.69KK24 pKa = 10.83RR25 pKa = 11.84MNQGRR30 pKa = 11.84RR31 pKa = 11.84WCFTWNNYY39 pKa = 9.2DD40 pKa = 4.0DD41 pKa = 4.71NVVTAIKK48 pKa = 10.25EE49 pKa = 4.03RR50 pKa = 11.84LEE52 pKa = 4.21SKK54 pKa = 10.68DD55 pKa = 2.86ILRR58 pKa = 11.84YY59 pKa = 9.54IVGFEE64 pKa = 3.6IGEE67 pKa = 4.3KK68 pKa = 10.36EE69 pKa = 4.2KK70 pKa = 10.47TPHH73 pKa = 5.19LQGYY77 pKa = 8.31FEE79 pKa = 4.37FTGQIARR86 pKa = 11.84RR87 pKa = 11.84PSEE90 pKa = 3.43MWDD93 pKa = 3.25NYY95 pKa = 10.58NLGRR99 pKa = 11.84FFKK102 pKa = 10.92CKK104 pKa = 8.74GTAEE108 pKa = 4.14EE109 pKa = 3.93NHH111 pKa = 6.76AYY113 pKa = 10.04CSKK116 pKa = 10.37SGNYY120 pKa = 8.19FQGGDD125 pKa = 3.08WSILKK130 pKa = 10.28NKK132 pKa = 9.48EE133 pKa = 3.77PLPIEE138 pKa = 4.11VLKK141 pKa = 11.31DD142 pKa = 2.97EE143 pKa = 5.21DD144 pKa = 4.67LYY146 pKa = 10.71PFQKK150 pKa = 10.58SIIDD154 pKa = 4.09IIDD157 pKa = 3.73GPVIKK162 pKa = 10.48GHH164 pKa = 5.66IHH166 pKa = 6.99WICDD170 pKa = 3.45LKK172 pKa = 10.84GQNGKK177 pKa = 7.95TEE179 pKa = 4.5LLRR182 pKa = 11.84KK183 pKa = 9.74LVVEE187 pKa = 4.53RR188 pKa = 11.84NLKK191 pKa = 8.25FTYY194 pKa = 9.98GGKK197 pKa = 9.91KK198 pKa = 9.94ADD200 pKa = 4.45IINLIYY206 pKa = 10.77NNKK209 pKa = 9.62QYY211 pKa = 11.64YY212 pKa = 8.24LTTKK216 pKa = 9.38NAAMVYY222 pKa = 9.86NLGRR226 pKa = 11.84SEE228 pKa = 4.79DD229 pKa = 3.96LEE231 pKa = 4.41KK232 pKa = 11.11VSYY235 pKa = 10.74TSMEE239 pKa = 4.11QISDD243 pKa = 3.65GLISNTKK250 pKa = 9.79YY251 pKa = 8.95EE252 pKa = 4.26TGCFVMNHH260 pKa = 4.9PHH262 pKa = 7.15IIVLANGMPSIRR274 pKa = 11.84SMTMSRR280 pKa = 11.84WIIYY284 pKa = 9.25KK285 pKa = 10.33INDD288 pKa = 3.34AKK290 pKa = 10.43EE291 pKa = 3.99LEE293 pKa = 4.37RR294 pKa = 11.84IEE296 pKa = 4.46YY297 pKa = 9.65EE298 pKa = 3.98SHH300 pKa = 6.86SNLGSPLDD308 pKa = 4.46DD309 pKa = 5.15LDD311 pKa = 3.64NN312 pKa = 4.35
MM1 pKa = 6.95PRR3 pKa = 11.84KK4 pKa = 9.13MEE6 pKa = 4.27SLEE9 pKa = 4.04SSGGGNSIPPPIKK22 pKa = 9.83EE23 pKa = 3.69KK24 pKa = 10.83RR25 pKa = 11.84MNQGRR30 pKa = 11.84RR31 pKa = 11.84WCFTWNNYY39 pKa = 9.2DD40 pKa = 4.0DD41 pKa = 4.71NVVTAIKK48 pKa = 10.25EE49 pKa = 4.03RR50 pKa = 11.84LEE52 pKa = 4.21SKK54 pKa = 10.68DD55 pKa = 2.86ILRR58 pKa = 11.84YY59 pKa = 9.54IVGFEE64 pKa = 3.6IGEE67 pKa = 4.3KK68 pKa = 10.36EE69 pKa = 4.2KK70 pKa = 10.47TPHH73 pKa = 5.19LQGYY77 pKa = 8.31FEE79 pKa = 4.37FTGQIARR86 pKa = 11.84RR87 pKa = 11.84PSEE90 pKa = 3.43MWDD93 pKa = 3.25NYY95 pKa = 10.58NLGRR99 pKa = 11.84FFKK102 pKa = 10.92CKK104 pKa = 8.74GTAEE108 pKa = 4.14EE109 pKa = 3.93NHH111 pKa = 6.76AYY113 pKa = 10.04CSKK116 pKa = 10.37SGNYY120 pKa = 8.19FQGGDD125 pKa = 3.08WSILKK130 pKa = 10.28NKK132 pKa = 9.48EE133 pKa = 3.77PLPIEE138 pKa = 4.11VLKK141 pKa = 11.31DD142 pKa = 2.97EE143 pKa = 5.21DD144 pKa = 4.67LYY146 pKa = 10.71PFQKK150 pKa = 10.58SIIDD154 pKa = 4.09IIDD157 pKa = 3.73GPVIKK162 pKa = 10.48GHH164 pKa = 5.66IHH166 pKa = 6.99WICDD170 pKa = 3.45LKK172 pKa = 10.84GQNGKK177 pKa = 7.95TEE179 pKa = 4.5LLRR182 pKa = 11.84KK183 pKa = 9.74LVVEE187 pKa = 4.53RR188 pKa = 11.84NLKK191 pKa = 8.25FTYY194 pKa = 9.98GGKK197 pKa = 9.91KK198 pKa = 9.94ADD200 pKa = 4.45IINLIYY206 pKa = 10.77NNKK209 pKa = 9.62QYY211 pKa = 11.64YY212 pKa = 8.24LTTKK216 pKa = 9.38NAAMVYY222 pKa = 9.86NLGRR226 pKa = 11.84SEE228 pKa = 4.79DD229 pKa = 3.96LEE231 pKa = 4.41KK232 pKa = 11.11VSYY235 pKa = 10.74TSMEE239 pKa = 4.11QISDD243 pKa = 3.65GLISNTKK250 pKa = 9.79YY251 pKa = 8.95EE252 pKa = 4.26TGCFVMNHH260 pKa = 4.9PHH262 pKa = 7.15IIVLANGMPSIRR274 pKa = 11.84SMTMSRR280 pKa = 11.84WIIYY284 pKa = 9.25KK285 pKa = 10.33INDD288 pKa = 3.34AKK290 pKa = 10.43EE291 pKa = 3.99LEE293 pKa = 4.37RR294 pKa = 11.84IEE296 pKa = 4.46YY297 pKa = 9.65EE298 pKa = 3.98SHH300 pKa = 6.86SNLGSPLDD308 pKa = 4.46DD309 pKa = 5.15LDD311 pKa = 3.64NN312 pKa = 4.35
Molecular weight: 36.03 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4UGW2|A0A0B4UGW2_9VIRU Putative capsid protein OS=Odonata-associated circular virus-16 OX=1592116 PE=4 SV=1
MM1 pKa = 6.85VRR3 pKa = 11.84SRR5 pKa = 11.84SRR7 pKa = 11.84SRR9 pKa = 11.84TSYY12 pKa = 9.8RR13 pKa = 11.84KK14 pKa = 8.93KK15 pKa = 9.44VSVKK19 pKa = 9.85RR20 pKa = 11.84RR21 pKa = 11.84VPRR24 pKa = 11.84TRR26 pKa = 11.84KK27 pKa = 9.81GVTVPVKK34 pKa = 10.55KK35 pKa = 10.39YY36 pKa = 7.12VQKK39 pKa = 10.74AIHH42 pKa = 5.86RR43 pKa = 11.84QIEE46 pKa = 4.38NKK48 pKa = 9.68CVQFNGAFTMVNYY61 pKa = 10.84ADD63 pKa = 4.6DD64 pKa = 3.53NTMRR68 pKa = 11.84GFALSPGTSISIAQGTGSADD88 pKa = 3.39RR89 pKa = 11.84TGNRR93 pKa = 11.84IKK95 pKa = 10.32IIKK98 pKa = 9.37GKK100 pKa = 9.7LVYY103 pKa = 10.31WMVPTAYY110 pKa = 10.29NGLLNPQPKK119 pKa = 9.71PLMVRR124 pKa = 11.84LWIGYY129 pKa = 9.54SKK131 pKa = 9.83TNPTLIPPAGDD142 pKa = 3.08QAVLFQNGNTASPPNGFINDD162 pKa = 3.65MLRR165 pKa = 11.84SVNKK169 pKa = 10.1DD170 pKa = 2.68KK171 pKa = 11.1FVIFRR176 pKa = 11.84DD177 pKa = 3.53IKK179 pKa = 10.83HH180 pKa = 5.93KK181 pKa = 10.75LGYY184 pKa = 10.27EE185 pKa = 4.4SITGTGFVAGSQYY198 pKa = 10.49FANNDD203 pKa = 3.17FKK205 pKa = 11.37FNIMRR210 pKa = 11.84TIDD213 pKa = 3.13ITKK216 pKa = 9.76YY217 pKa = 10.48LINNVIYY224 pKa = 10.64NDD226 pKa = 3.45TTAIPTSRR234 pKa = 11.84GLFCWVQIVYY244 pKa = 10.86ADD246 pKa = 3.78NNVTTGTIPANFNYY260 pKa = 9.68FVDD263 pKa = 4.5LTYY266 pKa = 11.34EE267 pKa = 4.05DD268 pKa = 3.87AA269 pKa = 5.76
MM1 pKa = 6.85VRR3 pKa = 11.84SRR5 pKa = 11.84SRR7 pKa = 11.84SRR9 pKa = 11.84TSYY12 pKa = 9.8RR13 pKa = 11.84KK14 pKa = 8.93KK15 pKa = 9.44VSVKK19 pKa = 9.85RR20 pKa = 11.84RR21 pKa = 11.84VPRR24 pKa = 11.84TRR26 pKa = 11.84KK27 pKa = 9.81GVTVPVKK34 pKa = 10.55KK35 pKa = 10.39YY36 pKa = 7.12VQKK39 pKa = 10.74AIHH42 pKa = 5.86RR43 pKa = 11.84QIEE46 pKa = 4.38NKK48 pKa = 9.68CVQFNGAFTMVNYY61 pKa = 10.84ADD63 pKa = 4.6DD64 pKa = 3.53NTMRR68 pKa = 11.84GFALSPGTSISIAQGTGSADD88 pKa = 3.39RR89 pKa = 11.84TGNRR93 pKa = 11.84IKK95 pKa = 10.32IIKK98 pKa = 9.37GKK100 pKa = 9.7LVYY103 pKa = 10.31WMVPTAYY110 pKa = 10.29NGLLNPQPKK119 pKa = 9.71PLMVRR124 pKa = 11.84LWIGYY129 pKa = 9.54SKK131 pKa = 9.83TNPTLIPPAGDD142 pKa = 3.08QAVLFQNGNTASPPNGFINDD162 pKa = 3.65MLRR165 pKa = 11.84SVNKK169 pKa = 10.1DD170 pKa = 2.68KK171 pKa = 11.1FVIFRR176 pKa = 11.84DD177 pKa = 3.53IKK179 pKa = 10.83HH180 pKa = 5.93KK181 pKa = 10.75LGYY184 pKa = 10.27EE185 pKa = 4.4SITGTGFVAGSQYY198 pKa = 10.49FANNDD203 pKa = 3.17FKK205 pKa = 11.37FNIMRR210 pKa = 11.84TIDD213 pKa = 3.13ITKK216 pKa = 9.76YY217 pKa = 10.48LINNVIYY224 pKa = 10.64NDD226 pKa = 3.45TTAIPTSRR234 pKa = 11.84GLFCWVQIVYY244 pKa = 10.86ADD246 pKa = 3.78NNVTTGTIPANFNYY260 pKa = 9.68FVDD263 pKa = 4.5LTYY266 pKa = 11.34EE267 pKa = 4.05DD268 pKa = 3.87AA269 pKa = 5.76
Molecular weight: 30.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
581 |
269 |
312 |
290.5 |
33.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.303 ± 1.07 | 1.205 ± 0.3 |
5.164 ± 0.215 | 4.819 ± 2.41 |
4.131 ± 0.698 | 7.401 ± 0.22 |
1.549 ± 0.524 | 8.434 ± 0.408 |
7.917 ± 0.556 | 6.368 ± 0.999 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.926 ± 0.211 | 7.917 ± 0.412 |
4.647 ± 0.363 | 2.926 ± 0.273 |
5.68 ± 0.658 | 6.196 ± 0.403 |
6.196 ± 1.531 | 5.68 ± 1.625 |
1.549 ± 0.282 | 4.991 ± 0.103 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
