
Methanosalsum zhilinae (strain DSM 4017 / NBRC 107636 / OCM 62 / WeN5) (Methanohalophilus zhilinae)
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Methanomicrobia; Methanosarcinales; Methanosarcinaceae; Methanosalsum; Methanosalsum zhilinae
Average proteome isoelectric point is 5.82
Get precalculated fractions of proteins
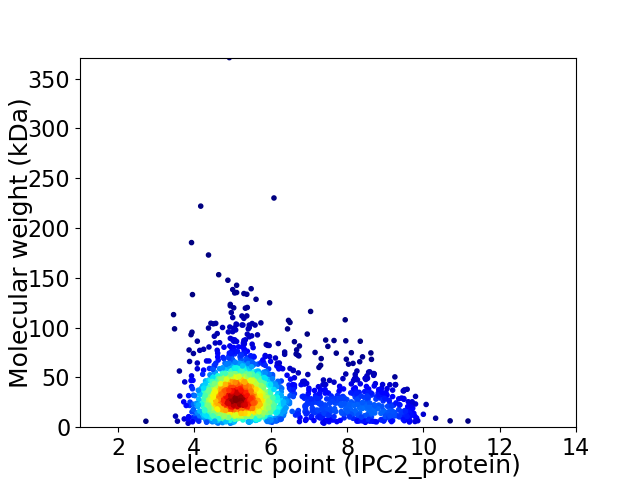
Virtual 2D-PAGE plot for 1972 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F7XLF0|F7XLF0_METZD Kynurenine formamidase OS=Methanosalsum zhilinae (strain DSM 4017 / NBRC 107636 / OCM 62 / WeN5) OX=679901 GN=Mzhil_0489 PE=4 SV=1
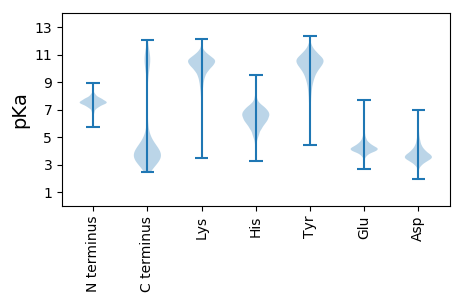
MM1 pKa = 7.59IKK3 pKa = 10.85GNILRR8 pKa = 11.84ILVILIALSLFGMHH22 pKa = 6.32SVASQDD28 pKa = 3.4SEE30 pKa = 3.84PRR32 pKa = 11.84EE33 pKa = 4.12FEE35 pKa = 4.16VLIQGFDD42 pKa = 3.64YY43 pKa = 11.13DD44 pKa = 3.65PSEE47 pKa = 4.96IIAHH51 pKa = 5.71VNDD54 pKa = 3.76TVVWTNMDD62 pKa = 3.92TVRR65 pKa = 11.84HH66 pKa = 5.21TVTADD71 pKa = 3.1EE72 pKa = 4.88FDD74 pKa = 5.31SGDD77 pKa = 4.03LDD79 pKa = 3.9QGDD82 pKa = 4.05SFNYY86 pKa = 9.42TFTEE90 pKa = 4.48PGTYY94 pKa = 9.78DD95 pKa = 3.47YY96 pKa = 11.85VCIYY100 pKa = 9.75HH101 pKa = 6.87PGMRR105 pKa = 11.84GTVIVEE111 pKa = 3.93AEE113 pKa = 3.9VDD115 pKa = 3.64YY116 pKa = 11.1DD117 pKa = 4.01VPPVADD123 pKa = 4.8FTANPTSGIAPLEE136 pKa = 3.91VSFTDD141 pKa = 3.38NSDD144 pKa = 3.51NAVEE148 pKa = 3.99YY149 pKa = 10.06HH150 pKa = 6.46WDD152 pKa = 3.6FGDD155 pKa = 3.94GQTSTEE161 pKa = 4.05VNPTHH166 pKa = 6.89TYY168 pKa = 9.48EE169 pKa = 4.46LPGAYY174 pKa = 8.13TVTLTVSNEE183 pKa = 3.72VGEE186 pKa = 4.69DD187 pKa = 3.26STEE190 pKa = 3.72EE191 pKa = 4.47FITVGHH197 pKa = 7.36PDD199 pKa = 4.88DD200 pKa = 5.91DD201 pKa = 4.44PAPAPGFGLIFAIAGLLVIASLVKK225 pKa = 10.5RR226 pKa = 11.84INEE229 pKa = 4.06KK230 pKa = 10.67
MM1 pKa = 7.59IKK3 pKa = 10.85GNILRR8 pKa = 11.84ILVILIALSLFGMHH22 pKa = 6.32SVASQDD28 pKa = 3.4SEE30 pKa = 3.84PRR32 pKa = 11.84EE33 pKa = 4.12FEE35 pKa = 4.16VLIQGFDD42 pKa = 3.64YY43 pKa = 11.13DD44 pKa = 3.65PSEE47 pKa = 4.96IIAHH51 pKa = 5.71VNDD54 pKa = 3.76TVVWTNMDD62 pKa = 3.92TVRR65 pKa = 11.84HH66 pKa = 5.21TVTADD71 pKa = 3.1EE72 pKa = 4.88FDD74 pKa = 5.31SGDD77 pKa = 4.03LDD79 pKa = 3.9QGDD82 pKa = 4.05SFNYY86 pKa = 9.42TFTEE90 pKa = 4.48PGTYY94 pKa = 9.78DD95 pKa = 3.47YY96 pKa = 11.85VCIYY100 pKa = 9.75HH101 pKa = 6.87PGMRR105 pKa = 11.84GTVIVEE111 pKa = 3.93AEE113 pKa = 3.9VDD115 pKa = 3.64YY116 pKa = 11.1DD117 pKa = 4.01VPPVADD123 pKa = 4.8FTANPTSGIAPLEE136 pKa = 3.91VSFTDD141 pKa = 3.38NSDD144 pKa = 3.51NAVEE148 pKa = 3.99YY149 pKa = 10.06HH150 pKa = 6.46WDD152 pKa = 3.6FGDD155 pKa = 3.94GQTSTEE161 pKa = 4.05VNPTHH166 pKa = 6.89TYY168 pKa = 9.48EE169 pKa = 4.46LPGAYY174 pKa = 8.13TVTLTVSNEE183 pKa = 3.72VGEE186 pKa = 4.69DD187 pKa = 3.26STEE190 pKa = 3.72EE191 pKa = 4.47FITVGHH197 pKa = 7.36PDD199 pKa = 4.88DD200 pKa = 5.91DD201 pKa = 4.44PAPAPGFGLIFAIAGLLVIASLVKK225 pKa = 10.5RR226 pKa = 11.84INEE229 pKa = 4.06KK230 pKa = 10.67
Molecular weight: 25.17 kDa
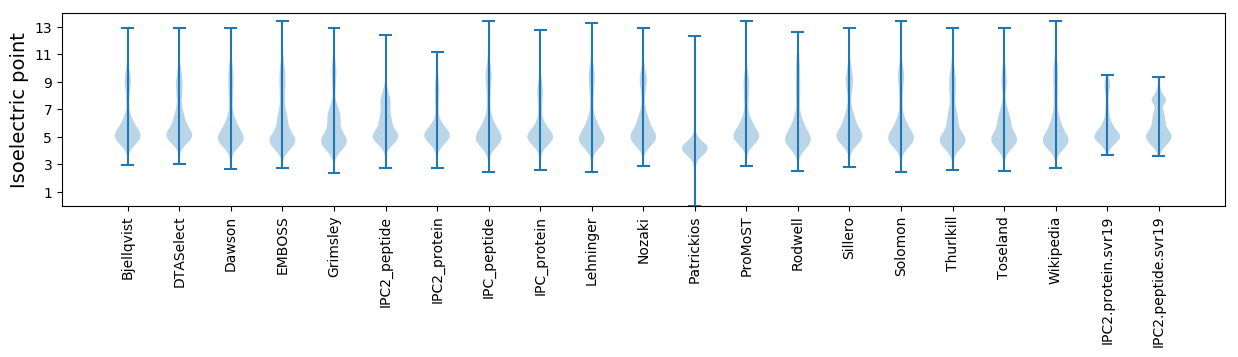
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F7XQ49|F7XQ49_METZD Tryptophan synthase alpha chain OS=Methanosalsum zhilinae (strain DSM 4017 / NBRC 107636 / OCM 62 / WeN5) OX=679901 GN=trpA PE=3 SV=1
MM1 pKa = 7.38SRR3 pKa = 11.84NTKK6 pKa = 7.84GQKK9 pKa = 8.18TRR11 pKa = 11.84LAKK14 pKa = 10.65AHH16 pKa = 5.79MQNQRR21 pKa = 11.84VPMWVIVKK29 pKa = 7.4TNRR32 pKa = 11.84SVVTHH37 pKa = 6.07PKK39 pKa = 8.16RR40 pKa = 11.84RR41 pKa = 11.84HH42 pKa = 3.92WRR44 pKa = 11.84RR45 pKa = 11.84SSLRR49 pKa = 11.84LKK51 pKa = 10.7
MM1 pKa = 7.38SRR3 pKa = 11.84NTKK6 pKa = 7.84GQKK9 pKa = 8.18TRR11 pKa = 11.84LAKK14 pKa = 10.65AHH16 pKa = 5.79MQNQRR21 pKa = 11.84VPMWVIVKK29 pKa = 7.4TNRR32 pKa = 11.84SVVTHH37 pKa = 6.07PKK39 pKa = 8.16RR40 pKa = 11.84RR41 pKa = 11.84HH42 pKa = 3.92WRR44 pKa = 11.84RR45 pKa = 11.84SSLRR49 pKa = 11.84LKK51 pKa = 10.7
Molecular weight: 6.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
610469 |
36 |
3261 |
309.6 |
34.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.514 ± 0.054 | 1.239 ± 0.028 |
6.319 ± 0.048 | 7.39 ± 0.052 |
3.84 ± 0.04 | 6.949 ± 0.058 |
2.017 ± 0.023 | 9.021 ± 0.05 |
6.203 ± 0.06 | 8.575 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.924 ± 0.028 | 4.588 ± 0.047 |
3.789 ± 0.034 | 2.674 ± 0.028 |
4.734 ± 0.046 | 6.926 ± 0.046 |
5.08 ± 0.04 | 6.863 ± 0.04 |
0.87 ± 0.021 | 3.484 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
