
Circovirus-like genome DHCV-5
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 8.28
Get precalculated fractions of proteins
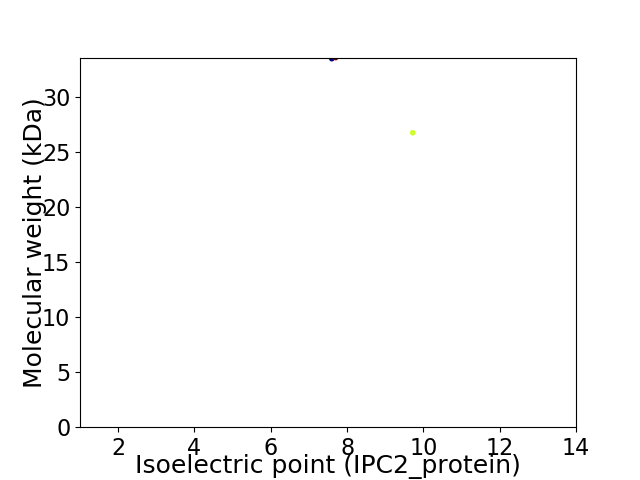
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A190WHE8|A0A190WHE8_9CIRC Uncharacterized protein OS=Circovirus-like genome DHCV-5 OX=1788454 PE=4 SV=1
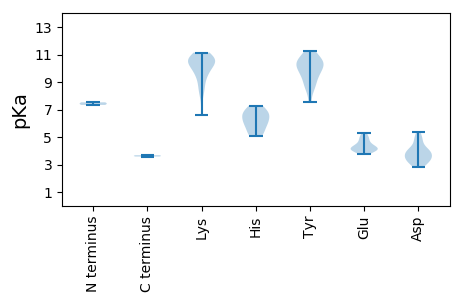
MM1 pKa = 7.53SSITMTQPVTKK12 pKa = 10.47ACDD15 pKa = 3.1KK16 pKa = 9.92TNRR19 pKa = 11.84SRR21 pKa = 11.84LTCFTIFDD29 pKa = 4.08MSIDD33 pKa = 3.94LQSLASRR40 pKa = 11.84FDD42 pKa = 3.6YY43 pKa = 10.15MAVGRR48 pKa = 11.84EE49 pKa = 4.3TCPSTQRR56 pKa = 11.84EE57 pKa = 4.37HH58 pKa = 6.19FQGFAYY64 pKa = 10.37CKK66 pKa = 8.34TAQRR70 pKa = 11.84FPWWHH75 pKa = 6.17KK76 pKa = 10.42RR77 pKa = 11.84LTPHH81 pKa = 6.73HH82 pKa = 6.89FEE84 pKa = 4.09FCFGSLEE91 pKa = 4.09QNEE94 pKa = 4.99RR95 pKa = 11.84YY96 pKa = 9.59CSKK99 pKa = 10.35QGQYY103 pKa = 9.75QEE105 pKa = 5.26FGIKK109 pKa = 10.17PMGDD113 pKa = 3.15GKK115 pKa = 10.78RR116 pKa = 11.84RR117 pKa = 11.84DD118 pKa = 3.53LSEE121 pKa = 4.03VCDD124 pKa = 4.77AIVTKK129 pKa = 10.25RR130 pKa = 11.84QKK132 pKa = 10.93LSDD135 pKa = 3.26IAIQFPTVFTQYY147 pKa = 11.25HH148 pKa = 5.73NGLTKK153 pKa = 10.71LEE155 pKa = 4.53AIITKK160 pKa = 9.5PYY162 pKa = 8.53EE163 pKa = 3.74HH164 pKa = 6.66HH165 pKa = 5.95TVRR168 pKa = 11.84GTWIQGVPGAGKK180 pKa = 8.55SHH182 pKa = 6.35YY183 pKa = 10.73ARR185 pKa = 11.84THH187 pKa = 5.09FQDD190 pKa = 4.2VYY192 pKa = 11.2IKK194 pKa = 10.67AQNKK198 pKa = 7.93WFDD201 pKa = 3.81GYY203 pKa = 9.42TGQHH207 pKa = 5.85TIILEE212 pKa = 4.28DD213 pKa = 4.02FDD215 pKa = 4.66SHH217 pKa = 5.57GHH219 pKa = 5.19YY220 pKa = 10.05LAHH223 pKa = 7.26LLKK226 pKa = 10.11IWSDD230 pKa = 2.87KK231 pKa = 9.66WSCSGEE237 pKa = 4.05IKK239 pKa = 10.7GGTVALQHH247 pKa = 6.83RR248 pKa = 11.84DD249 pKa = 3.47FVVTSNFSIDD259 pKa = 4.44EE260 pKa = 4.15IFTDD264 pKa = 3.89SKK266 pKa = 10.37FAEE269 pKa = 4.91PIKK272 pKa = 10.69RR273 pKa = 11.84RR274 pKa = 11.84FKK276 pKa = 10.82LITMDD281 pKa = 3.68TEE283 pKa = 4.72YY284 pKa = 10.54ISPDD288 pKa = 3.03SS289 pKa = 3.68
MM1 pKa = 7.53SSITMTQPVTKK12 pKa = 10.47ACDD15 pKa = 3.1KK16 pKa = 9.92TNRR19 pKa = 11.84SRR21 pKa = 11.84LTCFTIFDD29 pKa = 4.08MSIDD33 pKa = 3.94LQSLASRR40 pKa = 11.84FDD42 pKa = 3.6YY43 pKa = 10.15MAVGRR48 pKa = 11.84EE49 pKa = 4.3TCPSTQRR56 pKa = 11.84EE57 pKa = 4.37HH58 pKa = 6.19FQGFAYY64 pKa = 10.37CKK66 pKa = 8.34TAQRR70 pKa = 11.84FPWWHH75 pKa = 6.17KK76 pKa = 10.42RR77 pKa = 11.84LTPHH81 pKa = 6.73HH82 pKa = 6.89FEE84 pKa = 4.09FCFGSLEE91 pKa = 4.09QNEE94 pKa = 4.99RR95 pKa = 11.84YY96 pKa = 9.59CSKK99 pKa = 10.35QGQYY103 pKa = 9.75QEE105 pKa = 5.26FGIKK109 pKa = 10.17PMGDD113 pKa = 3.15GKK115 pKa = 10.78RR116 pKa = 11.84RR117 pKa = 11.84DD118 pKa = 3.53LSEE121 pKa = 4.03VCDD124 pKa = 4.77AIVTKK129 pKa = 10.25RR130 pKa = 11.84QKK132 pKa = 10.93LSDD135 pKa = 3.26IAIQFPTVFTQYY147 pKa = 11.25HH148 pKa = 5.73NGLTKK153 pKa = 10.71LEE155 pKa = 4.53AIITKK160 pKa = 9.5PYY162 pKa = 8.53EE163 pKa = 3.74HH164 pKa = 6.66HH165 pKa = 5.95TVRR168 pKa = 11.84GTWIQGVPGAGKK180 pKa = 8.55SHH182 pKa = 6.35YY183 pKa = 10.73ARR185 pKa = 11.84THH187 pKa = 5.09FQDD190 pKa = 4.2VYY192 pKa = 11.2IKK194 pKa = 10.67AQNKK198 pKa = 7.93WFDD201 pKa = 3.81GYY203 pKa = 9.42TGQHH207 pKa = 5.85TIILEE212 pKa = 4.28DD213 pKa = 4.02FDD215 pKa = 4.66SHH217 pKa = 5.57GHH219 pKa = 5.19YY220 pKa = 10.05LAHH223 pKa = 7.26LLKK226 pKa = 10.11IWSDD230 pKa = 2.87KK231 pKa = 9.66WSCSGEE237 pKa = 4.05IKK239 pKa = 10.7GGTVALQHH247 pKa = 6.83RR248 pKa = 11.84DD249 pKa = 3.47FVVTSNFSIDD259 pKa = 4.44EE260 pKa = 4.15IFTDD264 pKa = 3.89SKK266 pKa = 10.37FAEE269 pKa = 4.91PIKK272 pKa = 10.69RR273 pKa = 11.84RR274 pKa = 11.84FKK276 pKa = 10.82LITMDD281 pKa = 3.68TEE283 pKa = 4.72YY284 pKa = 10.54ISPDD288 pKa = 3.03SS289 pKa = 3.68
Molecular weight: 33.5 kDa
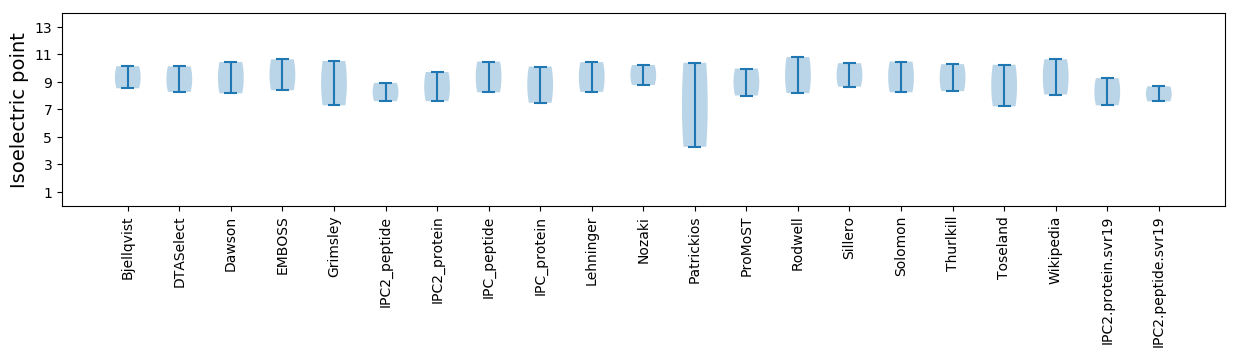
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A190WHE8|A0A190WHE8_9CIRC Uncharacterized protein OS=Circovirus-like genome DHCV-5 OX=1788454 PE=4 SV=1
MM1 pKa = 7.34TLLGKK6 pKa = 10.45RR7 pKa = 11.84KK8 pKa = 9.46GSGRR12 pKa = 11.84KK13 pKa = 6.64GTPRR17 pKa = 11.84KK18 pKa = 9.67RR19 pKa = 11.84PSIFARR25 pKa = 11.84RR26 pKa = 11.84PLTRR30 pKa = 11.84ARR32 pKa = 11.84NKK34 pKa = 10.21RR35 pKa = 11.84PRR37 pKa = 11.84TSKK40 pKa = 10.79LGALTTYY47 pKa = 10.2RR48 pKa = 11.84YY49 pKa = 10.34SRR51 pKa = 11.84WASTGQTHH59 pKa = 7.29DD60 pKa = 3.52VSTTSYY66 pKa = 10.51QGAATFTLDD75 pKa = 4.17DD76 pKa = 3.83VKK78 pKa = 11.14GYY80 pKa = 9.39TDD82 pKa = 4.0FTSLYY87 pKa = 8.59DD88 pKa = 3.42QFMITHH94 pKa = 6.37AQIHH98 pKa = 5.19VTLITNPDD106 pKa = 2.83SAYY109 pKa = 9.51PINGTGSSAVINPNNWFPKK128 pKa = 8.95LWYY131 pKa = 10.73VFDD134 pKa = 5.08SDD136 pKa = 5.35DD137 pKa = 4.94SSTLSLDD144 pKa = 3.94VIRR147 pKa = 11.84EE148 pKa = 4.1RR149 pKa = 11.84QGVKK153 pKa = 10.56YY154 pKa = 8.95KK155 pKa = 8.62TLRR158 pKa = 11.84PNSTMIINIKK168 pKa = 9.3PRR170 pKa = 11.84CLVQTYY176 pKa = 7.52RR177 pKa = 11.84TATSTGYY184 pKa = 10.73APRR187 pKa = 11.84RR188 pKa = 11.84MFLDD192 pKa = 3.15VATGYY197 pKa = 10.12NVPHH201 pKa = 6.74YY202 pKa = 10.22GLKK205 pKa = 10.45FCVDD209 pKa = 3.51TMGLDD214 pKa = 3.54PTDD217 pKa = 3.3KK218 pKa = 10.8FMVRR222 pKa = 11.84MEE224 pKa = 4.07TKK226 pKa = 9.77YY227 pKa = 8.69WLKK230 pKa = 10.8FKK232 pKa = 10.97GVLL235 pKa = 3.58
MM1 pKa = 7.34TLLGKK6 pKa = 10.45RR7 pKa = 11.84KK8 pKa = 9.46GSGRR12 pKa = 11.84KK13 pKa = 6.64GTPRR17 pKa = 11.84KK18 pKa = 9.67RR19 pKa = 11.84PSIFARR25 pKa = 11.84RR26 pKa = 11.84PLTRR30 pKa = 11.84ARR32 pKa = 11.84NKK34 pKa = 10.21RR35 pKa = 11.84PRR37 pKa = 11.84TSKK40 pKa = 10.79LGALTTYY47 pKa = 10.2RR48 pKa = 11.84YY49 pKa = 10.34SRR51 pKa = 11.84WASTGQTHH59 pKa = 7.29DD60 pKa = 3.52VSTTSYY66 pKa = 10.51QGAATFTLDD75 pKa = 4.17DD76 pKa = 3.83VKK78 pKa = 11.14GYY80 pKa = 9.39TDD82 pKa = 4.0FTSLYY87 pKa = 8.59DD88 pKa = 3.42QFMITHH94 pKa = 6.37AQIHH98 pKa = 5.19VTLITNPDD106 pKa = 2.83SAYY109 pKa = 9.51PINGTGSSAVINPNNWFPKK128 pKa = 8.95LWYY131 pKa = 10.73VFDD134 pKa = 5.08SDD136 pKa = 5.35DD137 pKa = 4.94SSTLSLDD144 pKa = 3.94VIRR147 pKa = 11.84EE148 pKa = 4.1RR149 pKa = 11.84QGVKK153 pKa = 10.56YY154 pKa = 8.95KK155 pKa = 8.62TLRR158 pKa = 11.84PNSTMIINIKK168 pKa = 9.3PRR170 pKa = 11.84CLVQTYY176 pKa = 7.52RR177 pKa = 11.84TATSTGYY184 pKa = 10.73APRR187 pKa = 11.84RR188 pKa = 11.84MFLDD192 pKa = 3.15VATGYY197 pKa = 10.12NVPHH201 pKa = 6.74YY202 pKa = 10.22GLKK205 pKa = 10.45FCVDD209 pKa = 3.51TMGLDD214 pKa = 3.54PTDD217 pKa = 3.3KK218 pKa = 10.8FMVRR222 pKa = 11.84MEE224 pKa = 4.07TKK226 pKa = 9.77YY227 pKa = 8.69WLKK230 pKa = 10.8FKK232 pKa = 10.97GVLL235 pKa = 3.58
Molecular weight: 26.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
524 |
235 |
289 |
262.0 |
30.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.962 ± 0.09 | 1.908 ± 0.658 |
6.107 ± 0.093 | 3.053 ± 1.371 |
5.725 ± 0.915 | 6.489 ± 0.199 |
3.435 ± 1.079 | 5.725 ± 0.915 |
6.87 ± 0.038 | 6.298 ± 0.848 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.481 ± 0.31 | 2.672 ± 0.721 |
4.389 ± 0.711 | 4.389 ± 1.143 |
6.87 ± 1.021 | 7.252 ± 0.011 |
10.305 ± 1.267 | 4.58 ± 0.592 |
1.908 ± 0.128 | 4.58 ± 0.592 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
