
Sphingopyxis sp. LC363
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Sphingopyxis; unclassified Sphingopyxis
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins
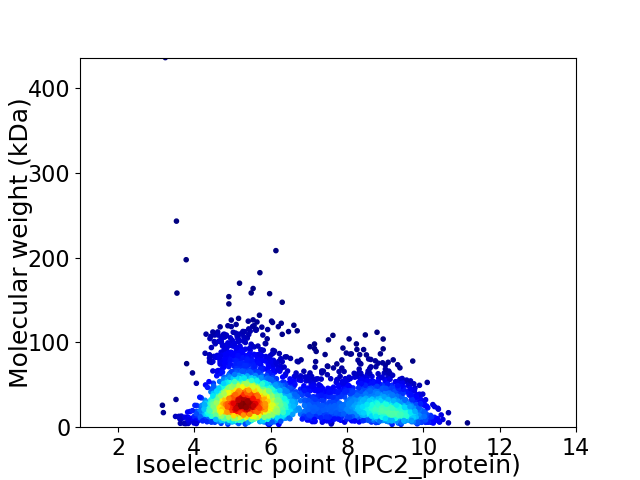
Virtual 2D-PAGE plot for 3880 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A095D7A4|A0A095D7A4_9SPHN Uncharacterized protein OS=Sphingopyxis sp. LC363 OX=1120705 GN=FG95_02616 PE=4 SV=1
MM1 pKa = 7.23LVWLAFTLAANPAIAQTTTTYY22 pKa = 11.49SNTTAGTINAATTCTALLVRR42 pKa = 11.84NFTVGASYY50 pKa = 9.78TVGDD54 pKa = 3.79VDD56 pKa = 5.31LGLLATHH63 pKa = 6.33SWRR66 pKa = 11.84GDD68 pKa = 3.32LRR70 pKa = 11.84VTLQSPAGTRR80 pKa = 11.84VQLVNGDD87 pKa = 3.53TGSIDD92 pKa = 3.27GNNFNVRR99 pKa = 11.84LDD101 pKa = 3.83DD102 pKa = 5.46DD103 pKa = 4.16AAQTVNTDD111 pKa = 3.29GNGTNHH117 pKa = 6.23SNSAPPYY124 pKa = 9.23QNVFSPNAPLSAFNGEE140 pKa = 3.99NSAGTWRR147 pKa = 11.84LEE149 pKa = 3.77ICDD152 pKa = 4.25QYY154 pKa = 11.7PSADD158 pKa = 3.04NGNFVRR164 pKa = 11.84ADD166 pKa = 3.86LYY168 pKa = 10.58LTGVPSSYY176 pKa = 11.58ADD178 pKa = 3.57LSLTKK183 pKa = 10.13SVSNAAPAFGASIDD197 pKa = 3.95YY198 pKa = 10.28VLSVTNASGSTLTATGVTVQDD219 pKa = 3.77NLPAGFNFTGASGFGSYY236 pKa = 10.47NSATGVWTVGSVPAGTTRR254 pKa = 11.84TLTISGTVAATAGASVTNSAEE275 pKa = 3.9IGASSAFDD283 pKa = 3.64FDD285 pKa = 4.05STPGNGAAGEE295 pKa = 4.5DD296 pKa = 4.26DD297 pKa = 3.9SDD299 pKa = 3.97SASFTVSGTRR309 pKa = 11.84VAGTPPALVCPVGTTVHH326 pKa = 6.84DD327 pKa = 4.06WDD329 pKa = 4.28SVTWPAGTTSAGAALTAIGSASFNIAVSGGSFLSNATYY367 pKa = 10.54GGQSPTRR374 pKa = 11.84QNVVTGGLTPAQYY387 pKa = 10.9SIFEE391 pKa = 4.42LVDD394 pKa = 3.73FTSQAGSVTTTITLPTAVPGAQFRR418 pKa = 11.84LFDD421 pKa = 3.32VDD423 pKa = 3.78YY424 pKa = 11.37ASGQFADD431 pKa = 3.84RR432 pKa = 11.84VTVTGTYY439 pKa = 10.31NGVAVTPILTNGTANYY455 pKa = 10.26VIGNSAYY462 pKa = 10.59GDD464 pKa = 3.68VLSADD469 pKa = 3.66GSADD473 pKa = 3.2GTVTVTFNAPVDD485 pKa = 4.66GITIEE490 pKa = 4.27YY491 pKa = 9.46GSHH494 pKa = 5.95SLAPANPGQQGMALHH509 pKa = 7.36DD510 pKa = 4.74IIFCRR515 pKa = 11.84PTANLVIAKK524 pKa = 7.96TSSVVSDD531 pKa = 4.03SVNGTANAKK540 pKa = 9.75AIPGARR546 pKa = 11.84MRR548 pKa = 11.84YY549 pKa = 9.37CILVTNNGSGTATGIAVADD568 pKa = 4.11ALPAGMSFIAGSLRR582 pKa = 11.84SGTSCAGATSVEE594 pKa = 4.07DD595 pKa = 4.47DD596 pKa = 3.78NANGADD602 pKa = 3.53EE603 pKa = 4.88SDD605 pKa = 3.49PFGASVAGTTVAATTATLLPAAAMAIAFEE634 pKa = 4.29VAIDD638 pKa = 3.46
MM1 pKa = 7.23LVWLAFTLAANPAIAQTTTTYY22 pKa = 11.49SNTTAGTINAATTCTALLVRR42 pKa = 11.84NFTVGASYY50 pKa = 9.78TVGDD54 pKa = 3.79VDD56 pKa = 5.31LGLLATHH63 pKa = 6.33SWRR66 pKa = 11.84GDD68 pKa = 3.32LRR70 pKa = 11.84VTLQSPAGTRR80 pKa = 11.84VQLVNGDD87 pKa = 3.53TGSIDD92 pKa = 3.27GNNFNVRR99 pKa = 11.84LDD101 pKa = 3.83DD102 pKa = 5.46DD103 pKa = 4.16AAQTVNTDD111 pKa = 3.29GNGTNHH117 pKa = 6.23SNSAPPYY124 pKa = 9.23QNVFSPNAPLSAFNGEE140 pKa = 3.99NSAGTWRR147 pKa = 11.84LEE149 pKa = 3.77ICDD152 pKa = 4.25QYY154 pKa = 11.7PSADD158 pKa = 3.04NGNFVRR164 pKa = 11.84ADD166 pKa = 3.86LYY168 pKa = 10.58LTGVPSSYY176 pKa = 11.58ADD178 pKa = 3.57LSLTKK183 pKa = 10.13SVSNAAPAFGASIDD197 pKa = 3.95YY198 pKa = 10.28VLSVTNASGSTLTATGVTVQDD219 pKa = 3.77NLPAGFNFTGASGFGSYY236 pKa = 10.47NSATGVWTVGSVPAGTTRR254 pKa = 11.84TLTISGTVAATAGASVTNSAEE275 pKa = 3.9IGASSAFDD283 pKa = 3.64FDD285 pKa = 4.05STPGNGAAGEE295 pKa = 4.5DD296 pKa = 4.26DD297 pKa = 3.9SDD299 pKa = 3.97SASFTVSGTRR309 pKa = 11.84VAGTPPALVCPVGTTVHH326 pKa = 6.84DD327 pKa = 4.06WDD329 pKa = 4.28SVTWPAGTTSAGAALTAIGSASFNIAVSGGSFLSNATYY367 pKa = 10.54GGQSPTRR374 pKa = 11.84QNVVTGGLTPAQYY387 pKa = 10.9SIFEE391 pKa = 4.42LVDD394 pKa = 3.73FTSQAGSVTTTITLPTAVPGAQFRR418 pKa = 11.84LFDD421 pKa = 3.32VDD423 pKa = 3.78YY424 pKa = 11.37ASGQFADD431 pKa = 3.84RR432 pKa = 11.84VTVTGTYY439 pKa = 10.31NGVAVTPILTNGTANYY455 pKa = 10.26VIGNSAYY462 pKa = 10.59GDD464 pKa = 3.68VLSADD469 pKa = 3.66GSADD473 pKa = 3.2GTVTVTFNAPVDD485 pKa = 4.66GITIEE490 pKa = 4.27YY491 pKa = 9.46GSHH494 pKa = 5.95SLAPANPGQQGMALHH509 pKa = 7.36DD510 pKa = 4.74IIFCRR515 pKa = 11.84PTANLVIAKK524 pKa = 7.96TSSVVSDD531 pKa = 4.03SVNGTANAKK540 pKa = 9.75AIPGARR546 pKa = 11.84MRR548 pKa = 11.84YY549 pKa = 9.37CILVTNNGSGTATGIAVADD568 pKa = 4.11ALPAGMSFIAGSLRR582 pKa = 11.84SGTSCAGATSVEE594 pKa = 4.07DD595 pKa = 4.47DD596 pKa = 3.78NANGADD602 pKa = 3.53EE603 pKa = 4.88SDD605 pKa = 3.49PFGASVAGTTVAATTATLLPAAAMAIAFEE634 pKa = 4.29VAIDD638 pKa = 3.46
Molecular weight: 64.03 kDa
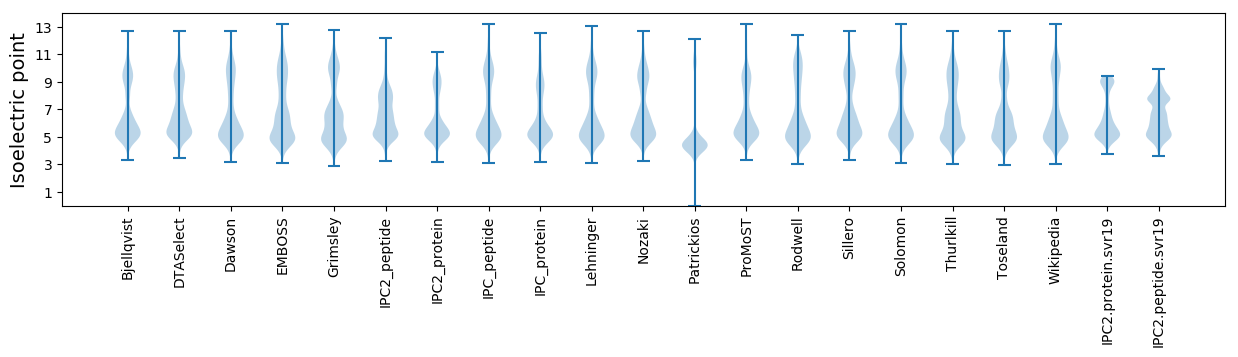
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A095B5L1|A0A095B5L1_9SPHN Uncharacterized protein OS=Sphingopyxis sp. LC363 OX=1120705 GN=FG95_01358 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.2QPSRR9 pKa = 11.84LVRR12 pKa = 11.84KK13 pKa = 9.21RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.57GFRR19 pKa = 11.84ARR21 pKa = 11.84KK22 pKa = 7.81ATVGGRR28 pKa = 11.84KK29 pKa = 9.05VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84AKK41 pKa = 10.7LSAA44 pKa = 4.0
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.2QPSRR9 pKa = 11.84LVRR12 pKa = 11.84KK13 pKa = 9.21RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.57GFRR19 pKa = 11.84ARR21 pKa = 11.84KK22 pKa = 7.81ATVGGRR28 pKa = 11.84KK29 pKa = 9.05VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84AKK41 pKa = 10.7LSAA44 pKa = 4.0
Molecular weight: 5.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1254710 |
29 |
4404 |
323.4 |
34.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.601 ± 0.065 | 0.742 ± 0.011 |
6.227 ± 0.03 | 5.427 ± 0.033 |
3.635 ± 0.024 | 9.029 ± 0.046 |
1.932 ± 0.02 | 5.092 ± 0.025 |
3.153 ± 0.032 | 9.688 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.404 ± 0.021 | 2.578 ± 0.029 |
5.324 ± 0.03 | 2.895 ± 0.02 |
7.23 ± 0.042 | 5.167 ± 0.028 |
5.152 ± 0.035 | 6.973 ± 0.028 |
1.49 ± 0.017 | 2.261 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
