
Kaistia algarum
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Kaistiaceae; Kaistia
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
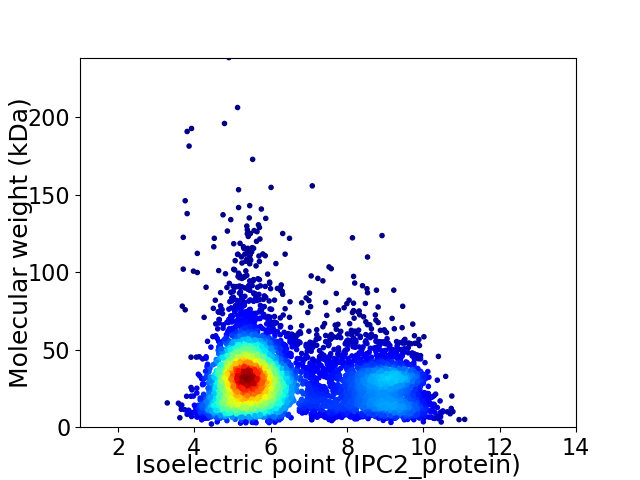
Virtual 2D-PAGE plot for 6004 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2S5TUP7|A0A2S5TUP7_9RHIZ Cell division coordinator CpoB OS=Kaistia algarum OX=2083279 GN=ygbF PE=3 SV=1
MM1 pKa = 7.32GVARR5 pKa = 11.84LLAATAFIAPSAALAQVQTGTCFAQALPSGGFAFSGSCQASSSSLIGLTGTGATGPNGSGADD67 pKa = 3.72LFYY70 pKa = 10.93SATDD74 pKa = 3.72GGPGATGPAFTFDD87 pKa = 4.61LLSGSQLLQQASAGNGLYY105 pKa = 10.63FSTTGGTGGIGGHH118 pKa = 6.49DD119 pKa = 3.31AVARR123 pKa = 11.84HH124 pKa = 6.11AGDD127 pKa = 3.84GGSGGDD133 pKa = 3.49GGVIIIDD140 pKa = 3.86ISATSAVSTTGDD152 pKa = 3.35NSDD155 pKa = 4.3AIYY158 pKa = 10.24AISSGGTGGQGGADD172 pKa = 3.28VDD174 pKa = 4.12GGLGDD179 pKa = 4.35GNGGPGGNGGDD190 pKa = 2.96ITVDD194 pKa = 3.0NDD196 pKa = 3.62GTLSTAGVYY205 pKa = 10.32SSAIFAHH212 pKa = 6.61SDD214 pKa = 2.99GGTGGAGAPAQSFFGNGGSGGVGGNSGADD243 pKa = 3.21VTARR247 pKa = 11.84NAGTASTSGDD257 pKa = 3.24NATAIVLQSVGGNGGDD273 pKa = 3.37GGGGQGAFYY282 pKa = 10.66SQGGNGGSGGNGEE295 pKa = 5.43DD296 pKa = 2.89IQFANRR302 pKa = 11.84GTIQTQGDD310 pKa = 3.67YY311 pKa = 11.39ADD313 pKa = 4.24AVNAYY318 pKa = 10.09SIGGGGGHH326 pKa = 6.87GGSAVTAGGLVGVAIGGSGGGGGDD350 pKa = 3.49SGSVALDD357 pKa = 3.22NFGTIIAAGTNAKK370 pKa = 10.15GMVGSSIGGGGGDD383 pKa = 3.66ANAVTNVTYY392 pKa = 10.88GPGAAWNFSFGGSGGMGGTGGAVVVDD418 pKa = 3.79NHH420 pKa = 6.73RR421 pKa = 11.84GGTIVTGAPTSNAPQDD437 pKa = 4.03PGNVPIPGDD446 pKa = 3.57HH447 pKa = 7.22ASGILAQSLGGGGGHH462 pKa = 7.1GAQTVSVSASAGYY475 pKa = 10.85ASFALGVALGGTGGAGGSGGTVSVTSDD502 pKa = 3.1GSITTHH508 pKa = 6.25ARR510 pKa = 11.84LADD513 pKa = 4.66GILAQSIGGGGGKK526 pKa = 10.28GGTSINVDD534 pKa = 3.08ASVGEE539 pKa = 4.21YY540 pKa = 10.46AGNLSIVLGGNGGNGGNGDD559 pKa = 4.01AVSVDD564 pKa = 3.42LGGAISTYY572 pKa = 10.23SSQSRR577 pKa = 11.84GIFAQSIGGGGGSGGNIINVDD598 pKa = 3.4AAVGSNAANLSVSLGGTGGGGGYY621 pKa = 10.08GSSVSVTTDD630 pKa = 2.7AGSTIATLGHH640 pKa = 6.2YY641 pKa = 10.27SDD643 pKa = 6.17GILAQSIGGGGGGGGSVNSYY663 pKa = 10.69AISAINGSGLAASAGVGVGGLGGSGGAGGDD693 pKa = 3.4VTVSHH698 pKa = 6.83GGAITTAGDD707 pKa = 3.41FATGLFAQSVGGGGGVGGSVFEE729 pKa = 4.37LSIAASLDD737 pKa = 3.37AQNAQGNALGGRR749 pKa = 11.84DD750 pKa = 3.37IAASIAVGGAGGAGNVGGDD769 pKa = 3.56VTVSLQSGSSIVTTGEE785 pKa = 3.71SAAGIIAQSIGGGGGAGGIAHH806 pKa = 6.92SFAVSTPVPTSPQRR820 pKa = 11.84LADD823 pKa = 4.49LLKK826 pKa = 9.88QWRR829 pKa = 11.84AMAEE833 pKa = 4.33KK834 pKa = 9.79IFSSQTSDD842 pKa = 3.33GPAGQYY848 pKa = 10.13QSGLQATISVGGNGGDD864 pKa = 3.34AGIGGNVIVNLASNASIQTSGSSSHH889 pKa = 6.07GVYY892 pKa = 10.53ALSIGGGGGIGGMANSDD909 pKa = 3.44GFAGFGEE916 pKa = 4.48FGLALSLGGNGGNDD930 pKa = 3.34NDD932 pKa = 4.37GGSVDD937 pKa = 4.0VVGSGPDD944 pKa = 3.23GTTAGIVTAGQLSHH958 pKa = 6.99GIAAQSVGGGGGSSASGGTGSTSDD982 pKa = 3.39HH983 pKa = 6.93SIPGLSRR990 pKa = 11.84QTVSMAIGGNAGAQGDD1006 pKa = 4.2GGSVRR1011 pKa = 11.84VIDD1014 pKa = 4.45APNIVTSGGGSDD1026 pKa = 5.14AIFAQSVGGGGGLGGAGSATGLIDD1050 pKa = 3.72ISLGGKK1056 pKa = 9.63GGAGGNGSTVRR1067 pKa = 11.84IKK1069 pKa = 10.94GDD1071 pKa = 3.94SILSTSGSASAGIFAQSVGGGGGAAGISNATLFGLQAGFDD1111 pKa = 3.86LRR1113 pKa = 11.84NGGIAGNGGQGGNVDD1128 pKa = 3.75VTWNGRR1134 pKa = 11.84IQTTGAVSIGIFAQSIGGGGGAVAANDD1161 pKa = 3.53LTIVGGAVPFYY1172 pKa = 9.64SLPGGGVGNGGTVTVGDD1189 pKa = 4.51LANDD1193 pKa = 4.12PLRR1196 pKa = 11.84LSTSGDD1202 pKa = 3.48GAHH1205 pKa = 7.11GIVAQSIGGGGVGLFADD1222 pKa = 4.07QPTEE1226 pKa = 3.87LFDD1229 pKa = 4.89VIWTAGGTPNGMGGSVIVTLNGSVATSGANAYY1261 pKa = 10.27GIVAQSLTNATVLFGTDD1278 pKa = 3.48GIEE1281 pKa = 4.07IVQEE1285 pKa = 4.15PTPSAQGAVQVILQGSVTTQGVGAHH1310 pKa = 7.21GIATYY1315 pKa = 10.52MNTTQAGNPVVQATGTVNVSGAGAWGIYY1343 pKa = 5.93TTNGQDD1349 pKa = 3.39GFGAPSTEE1357 pKa = 4.17AFTTDD1362 pKa = 2.8ISIQAGGTINATGAAAGAINMADD1385 pKa = 3.25WNGEE1389 pKa = 3.65ATADD1393 pKa = 2.84IWGALNAPDD1402 pKa = 5.2AVALQSNAAGLLNVHH1417 pKa = 6.34SGGVVLGDD1425 pKa = 3.47IVATGGDD1432 pKa = 4.31FILLNEE1438 pKa = 4.19GSITGSVSGVSTYY1451 pKa = 11.03AIGGGSHH1458 pKa = 7.2FLRR1461 pKa = 11.84FDD1463 pKa = 3.24PTATLGSDD1471 pKa = 4.18SIAVQQLEE1479 pKa = 4.45VPSNSIHH1486 pKa = 6.58PVLIALPNGNVTATQIIEE1504 pKa = 4.24THH1506 pKa = 6.2GGADD1510 pKa = 4.02LTPVPADD1517 pKa = 3.29QVGSFGLSSGTIATQYY1533 pKa = 11.26DD1534 pKa = 3.9YY1535 pKa = 11.74DD1536 pKa = 4.16FTLSTATIADD1546 pKa = 3.36VSIDD1550 pKa = 3.5FTRR1553 pKa = 11.84GGFSGNTQQLASAANNQLLGWAADD1577 pKa = 3.78APTSPSALYY1586 pKa = 9.61TLLLNASNATTSAEE1600 pKa = 3.74LSQYY1604 pKa = 10.04LQNLDD1609 pKa = 3.19ATASYY1614 pKa = 11.02AAVQQSAMAASAAHH1628 pKa = 5.67TTMQSCGNPVGLYY1641 pKa = 10.55SLIDD1645 pKa = 3.42QSPCNWGKK1653 pKa = 8.58ATYY1656 pKa = 10.51AVTTLRR1662 pKa = 11.84DD1663 pKa = 3.02GDD1665 pKa = 3.86QRR1667 pKa = 11.84DD1668 pKa = 3.76TTTGISIGRR1677 pKa = 11.84QDD1679 pKa = 3.55EE1680 pKa = 4.37ASDD1683 pKa = 3.54NVYY1686 pKa = 10.89VGGSFAYY1693 pKa = 10.07DD1694 pKa = 2.96WTSFDD1699 pKa = 4.02GVGSSSDD1706 pKa = 3.4GNRR1709 pKa = 11.84VSLGAIAKK1717 pKa = 8.61YY1718 pKa = 10.8VEE1720 pKa = 4.27GPVYY1724 pKa = 10.75ASASVVGSYY1733 pKa = 9.56GWADD1737 pKa = 3.4GSRR1740 pKa = 11.84YY1741 pKa = 10.17SSLSWVNGVATADD1754 pKa = 3.54QEE1756 pKa = 4.55TWALTGRR1763 pKa = 11.84LRR1765 pKa = 11.84AGYY1768 pKa = 9.8VFDD1771 pKa = 3.84VGAVEE1776 pKa = 5.71LMPLVDD1782 pKa = 5.79FDD1784 pKa = 3.89VQGIFDD1790 pKa = 3.39QGYY1793 pKa = 7.29TEE1795 pKa = 5.07QGLDD1799 pKa = 3.67DD1800 pKa = 4.98LALQVSNSNNVLFDD1814 pKa = 3.75LRR1816 pKa = 11.84PALRR1820 pKa = 11.84IGGTTKK1826 pKa = 10.57VNDD1829 pKa = 3.99AYY1831 pKa = 9.53ITNHH1835 pKa = 5.29VEE1837 pKa = 3.45IGALFALNEE1846 pKa = 4.1GSVDD1850 pKa = 3.33VSLPNGPNPTASVPLALEE1868 pKa = 4.46RR1869 pKa = 11.84DD1870 pKa = 3.77DD1871 pKa = 4.95VMATIALGSSIDD1883 pKa = 3.16WGAYY1887 pKa = 6.96EE1888 pKa = 6.11LRR1890 pKa = 11.84LQYY1893 pKa = 10.62EE1894 pKa = 4.53GALGDD1899 pKa = 3.94TTVSQAGSVKK1909 pKa = 10.16FAIKK1913 pKa = 10.25FF1914 pKa = 3.5
MM1 pKa = 7.32GVARR5 pKa = 11.84LLAATAFIAPSAALAQVQTGTCFAQALPSGGFAFSGSCQASSSSLIGLTGTGATGPNGSGADD67 pKa = 3.72LFYY70 pKa = 10.93SATDD74 pKa = 3.72GGPGATGPAFTFDD87 pKa = 4.61LLSGSQLLQQASAGNGLYY105 pKa = 10.63FSTTGGTGGIGGHH118 pKa = 6.49DD119 pKa = 3.31AVARR123 pKa = 11.84HH124 pKa = 6.11AGDD127 pKa = 3.84GGSGGDD133 pKa = 3.49GGVIIIDD140 pKa = 3.86ISATSAVSTTGDD152 pKa = 3.35NSDD155 pKa = 4.3AIYY158 pKa = 10.24AISSGGTGGQGGADD172 pKa = 3.28VDD174 pKa = 4.12GGLGDD179 pKa = 4.35GNGGPGGNGGDD190 pKa = 2.96ITVDD194 pKa = 3.0NDD196 pKa = 3.62GTLSTAGVYY205 pKa = 10.32SSAIFAHH212 pKa = 6.61SDD214 pKa = 2.99GGTGGAGAPAQSFFGNGGSGGVGGNSGADD243 pKa = 3.21VTARR247 pKa = 11.84NAGTASTSGDD257 pKa = 3.24NATAIVLQSVGGNGGDD273 pKa = 3.37GGGGQGAFYY282 pKa = 10.66SQGGNGGSGGNGEE295 pKa = 5.43DD296 pKa = 2.89IQFANRR302 pKa = 11.84GTIQTQGDD310 pKa = 3.67YY311 pKa = 11.39ADD313 pKa = 4.24AVNAYY318 pKa = 10.09SIGGGGGHH326 pKa = 6.87GGSAVTAGGLVGVAIGGSGGGGGDD350 pKa = 3.49SGSVALDD357 pKa = 3.22NFGTIIAAGTNAKK370 pKa = 10.15GMVGSSIGGGGGDD383 pKa = 3.66ANAVTNVTYY392 pKa = 10.88GPGAAWNFSFGGSGGMGGTGGAVVVDD418 pKa = 3.79NHH420 pKa = 6.73RR421 pKa = 11.84GGTIVTGAPTSNAPQDD437 pKa = 4.03PGNVPIPGDD446 pKa = 3.57HH447 pKa = 7.22ASGILAQSLGGGGGHH462 pKa = 7.1GAQTVSVSASAGYY475 pKa = 10.85ASFALGVALGGTGGAGGSGGTVSVTSDD502 pKa = 3.1GSITTHH508 pKa = 6.25ARR510 pKa = 11.84LADD513 pKa = 4.66GILAQSIGGGGGKK526 pKa = 10.28GGTSINVDD534 pKa = 3.08ASVGEE539 pKa = 4.21YY540 pKa = 10.46AGNLSIVLGGNGGNGGNGDD559 pKa = 4.01AVSVDD564 pKa = 3.42LGGAISTYY572 pKa = 10.23SSQSRR577 pKa = 11.84GIFAQSIGGGGGSGGNIINVDD598 pKa = 3.4AAVGSNAANLSVSLGGTGGGGGYY621 pKa = 10.08GSSVSVTTDD630 pKa = 2.7AGSTIATLGHH640 pKa = 6.2YY641 pKa = 10.27SDD643 pKa = 6.17GILAQSIGGGGGGGGSVNSYY663 pKa = 10.69AISAINGSGLAASAGVGVGGLGGSGGAGGDD693 pKa = 3.4VTVSHH698 pKa = 6.83GGAITTAGDD707 pKa = 3.41FATGLFAQSVGGGGGVGGSVFEE729 pKa = 4.37LSIAASLDD737 pKa = 3.37AQNAQGNALGGRR749 pKa = 11.84DD750 pKa = 3.37IAASIAVGGAGGAGNVGGDD769 pKa = 3.56VTVSLQSGSSIVTTGEE785 pKa = 3.71SAAGIIAQSIGGGGGAGGIAHH806 pKa = 6.92SFAVSTPVPTSPQRR820 pKa = 11.84LADD823 pKa = 4.49LLKK826 pKa = 9.88QWRR829 pKa = 11.84AMAEE833 pKa = 4.33KK834 pKa = 9.79IFSSQTSDD842 pKa = 3.33GPAGQYY848 pKa = 10.13QSGLQATISVGGNGGDD864 pKa = 3.34AGIGGNVIVNLASNASIQTSGSSSHH889 pKa = 6.07GVYY892 pKa = 10.53ALSIGGGGGIGGMANSDD909 pKa = 3.44GFAGFGEE916 pKa = 4.48FGLALSLGGNGGNDD930 pKa = 3.34NDD932 pKa = 4.37GGSVDD937 pKa = 4.0VVGSGPDD944 pKa = 3.23GTTAGIVTAGQLSHH958 pKa = 6.99GIAAQSVGGGGGSSASGGTGSTSDD982 pKa = 3.39HH983 pKa = 6.93SIPGLSRR990 pKa = 11.84QTVSMAIGGNAGAQGDD1006 pKa = 4.2GGSVRR1011 pKa = 11.84VIDD1014 pKa = 4.45APNIVTSGGGSDD1026 pKa = 5.14AIFAQSVGGGGGLGGAGSATGLIDD1050 pKa = 3.72ISLGGKK1056 pKa = 9.63GGAGGNGSTVRR1067 pKa = 11.84IKK1069 pKa = 10.94GDD1071 pKa = 3.94SILSTSGSASAGIFAQSVGGGGGAAGISNATLFGLQAGFDD1111 pKa = 3.86LRR1113 pKa = 11.84NGGIAGNGGQGGNVDD1128 pKa = 3.75VTWNGRR1134 pKa = 11.84IQTTGAVSIGIFAQSIGGGGGAVAANDD1161 pKa = 3.53LTIVGGAVPFYY1172 pKa = 9.64SLPGGGVGNGGTVTVGDD1189 pKa = 4.51LANDD1193 pKa = 4.12PLRR1196 pKa = 11.84LSTSGDD1202 pKa = 3.48GAHH1205 pKa = 7.11GIVAQSIGGGGVGLFADD1222 pKa = 4.07QPTEE1226 pKa = 3.87LFDD1229 pKa = 4.89VIWTAGGTPNGMGGSVIVTLNGSVATSGANAYY1261 pKa = 10.27GIVAQSLTNATVLFGTDD1278 pKa = 3.48GIEE1281 pKa = 4.07IVQEE1285 pKa = 4.15PTPSAQGAVQVILQGSVTTQGVGAHH1310 pKa = 7.21GIATYY1315 pKa = 10.52MNTTQAGNPVVQATGTVNVSGAGAWGIYY1343 pKa = 5.93TTNGQDD1349 pKa = 3.39GFGAPSTEE1357 pKa = 4.17AFTTDD1362 pKa = 2.8ISIQAGGTINATGAAAGAINMADD1385 pKa = 3.25WNGEE1389 pKa = 3.65ATADD1393 pKa = 2.84IWGALNAPDD1402 pKa = 5.2AVALQSNAAGLLNVHH1417 pKa = 6.34SGGVVLGDD1425 pKa = 3.47IVATGGDD1432 pKa = 4.31FILLNEE1438 pKa = 4.19GSITGSVSGVSTYY1451 pKa = 11.03AIGGGSHH1458 pKa = 7.2FLRR1461 pKa = 11.84FDD1463 pKa = 3.24PTATLGSDD1471 pKa = 4.18SIAVQQLEE1479 pKa = 4.45VPSNSIHH1486 pKa = 6.58PVLIALPNGNVTATQIIEE1504 pKa = 4.24THH1506 pKa = 6.2GGADD1510 pKa = 4.02LTPVPADD1517 pKa = 3.29QVGSFGLSSGTIATQYY1533 pKa = 11.26DD1534 pKa = 3.9YY1535 pKa = 11.74DD1536 pKa = 4.16FTLSTATIADD1546 pKa = 3.36VSIDD1550 pKa = 3.5FTRR1553 pKa = 11.84GGFSGNTQQLASAANNQLLGWAADD1577 pKa = 3.78APTSPSALYY1586 pKa = 9.61TLLLNASNATTSAEE1600 pKa = 3.74LSQYY1604 pKa = 10.04LQNLDD1609 pKa = 3.19ATASYY1614 pKa = 11.02AAVQQSAMAASAAHH1628 pKa = 5.67TTMQSCGNPVGLYY1641 pKa = 10.55SLIDD1645 pKa = 3.42QSPCNWGKK1653 pKa = 8.58ATYY1656 pKa = 10.51AVTTLRR1662 pKa = 11.84DD1663 pKa = 3.02GDD1665 pKa = 3.86QRR1667 pKa = 11.84DD1668 pKa = 3.76TTTGISIGRR1677 pKa = 11.84QDD1679 pKa = 3.55EE1680 pKa = 4.37ASDD1683 pKa = 3.54NVYY1686 pKa = 10.89VGGSFAYY1693 pKa = 10.07DD1694 pKa = 2.96WTSFDD1699 pKa = 4.02GVGSSSDD1706 pKa = 3.4GNRR1709 pKa = 11.84VSLGAIAKK1717 pKa = 8.61YY1718 pKa = 10.8VEE1720 pKa = 4.27GPVYY1724 pKa = 10.75ASASVVGSYY1733 pKa = 9.56GWADD1737 pKa = 3.4GSRR1740 pKa = 11.84YY1741 pKa = 10.17SSLSWVNGVATADD1754 pKa = 3.54QEE1756 pKa = 4.55TWALTGRR1763 pKa = 11.84LRR1765 pKa = 11.84AGYY1768 pKa = 9.8VFDD1771 pKa = 3.84VGAVEE1776 pKa = 5.71LMPLVDD1782 pKa = 5.79FDD1784 pKa = 3.89VQGIFDD1790 pKa = 3.39QGYY1793 pKa = 7.29TEE1795 pKa = 5.07QGLDD1799 pKa = 3.67DD1800 pKa = 4.98LALQVSNSNNVLFDD1814 pKa = 3.75LRR1816 pKa = 11.84PALRR1820 pKa = 11.84IGGTTKK1826 pKa = 10.57VNDD1829 pKa = 3.99AYY1831 pKa = 9.53ITNHH1835 pKa = 5.29VEE1837 pKa = 3.45IGALFALNEE1846 pKa = 4.1GSVDD1850 pKa = 3.33VSLPNGPNPTASVPLALEE1868 pKa = 4.46RR1869 pKa = 11.84DD1870 pKa = 3.77DD1871 pKa = 4.95VMATIALGSSIDD1883 pKa = 3.16WGAYY1887 pKa = 6.96EE1888 pKa = 6.11LRR1890 pKa = 11.84LQYY1893 pKa = 10.62EE1894 pKa = 4.53GALGDD1899 pKa = 3.94TTVSQAGSVKK1909 pKa = 10.16FAIKK1913 pKa = 10.25FF1914 pKa = 3.5
Molecular weight: 181.48 kDa
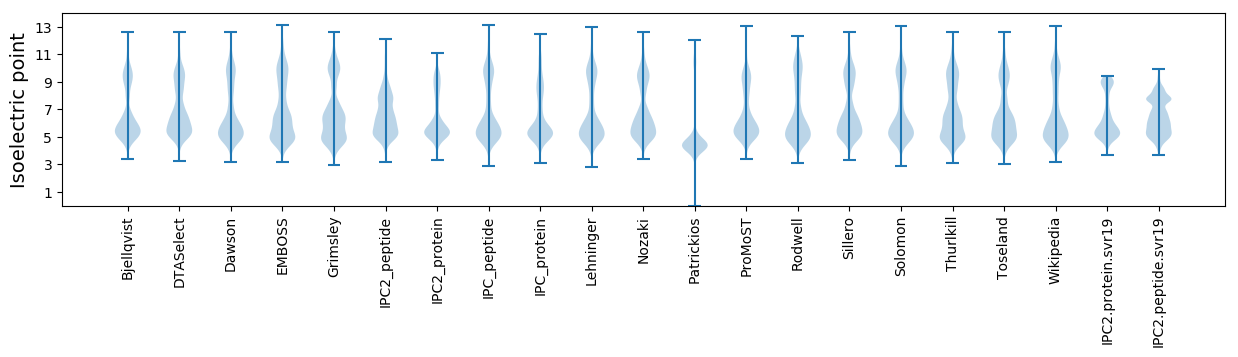
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2S5TS36|A0A2S5TS36_9RHIZ Fatty acid desaturase OS=Kaistia algarum OX=2083279 GN=C3941_22845 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.74LVRR12 pKa = 11.84TRR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.65GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.31VIAARR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84KK40 pKa = 9.55KK41 pKa = 9.66LTAA44 pKa = 4.17
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.74LVRR12 pKa = 11.84TRR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.65GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.31VIAARR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84KK40 pKa = 9.55KK41 pKa = 9.66LTAA44 pKa = 4.17
Molecular weight: 5.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1794267 |
20 |
2242 |
298.8 |
32.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.395 ± 0.051 | 0.741 ± 0.01 |
5.635 ± 0.028 | 5.565 ± 0.031 |
3.898 ± 0.021 | 8.618 ± 0.038 |
1.971 ± 0.015 | 5.74 ± 0.032 |
3.409 ± 0.03 | 10.19 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.327 ± 0.013 | 2.687 ± 0.021 |
5.137 ± 0.026 | 3.051 ± 0.021 |
6.68 ± 0.035 | 5.652 ± 0.024 |
5.205 ± 0.022 | 7.453 ± 0.03 |
1.323 ± 0.012 | 2.32 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
