
Sphingomonas sp. Root241
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Sphingomonas; unclassified Sphingomonas
Average proteome isoelectric point is 6.56
Get precalculated fractions of proteins
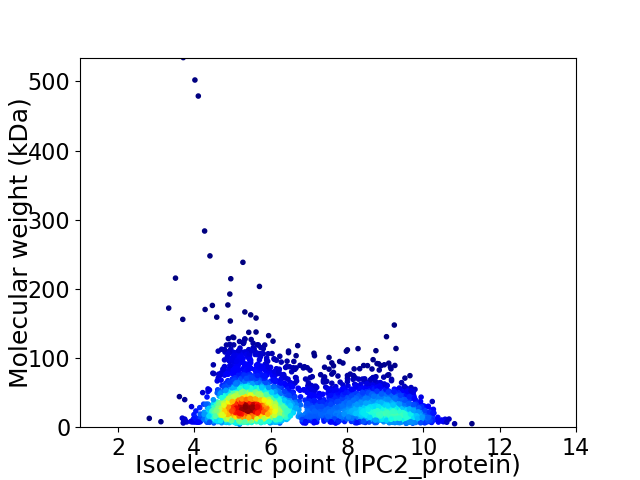
Virtual 2D-PAGE plot for 3715 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q8XKD7|A0A0Q8XKD7_9SPHN Uncharacterized protein OS=Sphingomonas sp. Root241 OX=1736501 GN=ASE13_09965 PE=4 SV=1
MM1 pKa = 7.75PAPAFINEE9 pKa = 3.72IHH11 pKa = 6.67YY12 pKa = 11.02DD13 pKa = 3.48NAGADD18 pKa = 3.16SGEE21 pKa = 4.29FIEE24 pKa = 4.58IAGIAGTDD32 pKa = 3.36LTGWKK37 pKa = 9.47IVLYY41 pKa = 10.42NGANGASYY49 pKa = 10.92NPVMTLSGVIANQQNGFGTISVAYY73 pKa = 7.04PTDD76 pKa = 5.04GIQNGSPDD84 pKa = 3.6AVALVDD90 pKa = 3.72ASNNVVQFLSYY101 pKa = 10.23EE102 pKa = 4.25GSFTATNGPAAGLTSVNVGVSEE124 pKa = 4.59NGAQSGTSIGLVGSGATYY142 pKa = 11.04EE143 pKa = 4.1DD144 pKa = 4.15FHH146 pKa = 6.57WALINGSTAGGVNAGQSFNGVVPPQPGTLSIADD179 pKa = 3.55ATTVEE184 pKa = 4.95GNSGTHH190 pKa = 6.12EE191 pKa = 3.9IVFTVTRR198 pKa = 11.84ADD200 pKa = 3.54GSAGAVSATWTVAFGSADD218 pKa = 3.41AADD221 pKa = 5.0FGAGFTATGTVSFADD236 pKa = 3.79GATTAEE242 pKa = 3.84IRR244 pKa = 11.84LPVQGDD250 pKa = 3.56TAFEE254 pKa = 4.53SNDD257 pKa = 2.94GFTVRR262 pKa = 11.84LSAPQGGVALGDD274 pKa = 3.46AVAAGTITNDD284 pKa = 3.16AAAPPAPPANVFINEE299 pKa = 3.67IHH301 pKa = 6.64YY302 pKa = 11.22DD303 pKa = 3.62NAGTDD308 pKa = 3.05AGEE311 pKa = 4.54AIEE314 pKa = 4.2IAGAAGTDD322 pKa = 3.33LTGYY326 pKa = 10.95KK327 pKa = 10.2LVFYY331 pKa = 10.45NGSNTPGAAPVYY343 pKa = 8.55DD344 pKa = 3.88TLALSGVIDD353 pKa = 4.37DD354 pKa = 4.55EE355 pKa = 5.44SNGFGALGFLRR366 pKa = 11.84AGIQNGAADD375 pKa = 4.78GVALIAPDD383 pKa = 3.82GSVVQLLSYY392 pKa = 10.22EE393 pKa = 4.46GSFTAAAGTPAAGTTSTDD411 pKa = 2.58IGVTEE416 pKa = 4.77EE417 pKa = 3.97PAPAAGLSLQLKK429 pKa = 10.11GSGSSAADD437 pKa = 3.63FAWADD442 pKa = 3.4ASDD445 pKa = 4.28DD446 pKa = 4.11SFGSLNAGQSFLSATGTGHH465 pKa = 7.31LRR467 pKa = 11.84IGDD470 pKa = 3.69ARR472 pKa = 11.84VVEE475 pKa = 4.64GDD477 pKa = 3.25SGTTNLVFTVNRR489 pKa = 11.84AGGTALAATVDD500 pKa = 3.81YY501 pKa = 10.91AVNLDD506 pKa = 3.74GTATLADD513 pKa = 4.51LAPGAVLSGTLSFAPGEE530 pKa = 3.96FSKK533 pKa = 11.09QIVVAVNGDD542 pKa = 3.77LVGEE546 pKa = 4.18PNEE549 pKa = 4.18TLSVSLGATTGNVVIDD565 pKa = 4.1DD566 pKa = 4.05AAATGTITNDD576 pKa = 3.17DD577 pKa = 4.98PIALTISQIQGAGHH591 pKa = 5.89VSAYY595 pKa = 10.14AGQVVITTGIVTAVDD610 pKa = 3.38TNGFYY615 pKa = 10.84LQSAVGDD622 pKa = 3.77GDD624 pKa = 4.29AATSDD629 pKa = 3.29AVFVFTSTAPGVVMGDD645 pKa = 3.58GVSVRR650 pKa = 11.84GSVAEE655 pKa = 4.23FQGSTASLSLTEE667 pKa = 4.35IVAPTVTVEE676 pKa = 3.88THH678 pKa = 6.27GNALPTALLIGTGGILPPSEE698 pKa = 5.17VIDD701 pKa = 4.65DD702 pKa = 5.03DD703 pKa = 5.02GLTSYY708 pKa = 11.34DD709 pKa = 4.01PLHH712 pKa = 7.16DD713 pKa = 5.28GVDD716 pKa = 3.99FWEE719 pKa = 4.5SLEE722 pKa = 3.93GMRR725 pKa = 11.84VTLDD729 pKa = 3.19TPQAVSNTTSFGEE742 pKa = 3.97TDD744 pKa = 3.56VVVSHH749 pKa = 6.65GNGASGINDD758 pKa = 3.41RR759 pKa = 11.84GGITISGDD767 pKa = 3.09GSGVPDD773 pKa = 3.85YY774 pKa = 11.02NPEE777 pKa = 4.5KK778 pKa = 10.6IQIDD782 pKa = 3.88DD783 pKa = 3.97DD784 pKa = 4.42SGIFAGFTPGYY795 pKa = 9.03TIGDD799 pKa = 3.71QLSSVTGVVNYY810 pKa = 10.68AFDD813 pKa = 3.98AYY815 pKa = 10.06EE816 pKa = 4.08VLVTEE821 pKa = 5.03AVTVTQDD828 pKa = 2.95TTLTRR833 pKa = 11.84EE834 pKa = 3.85QTALHH839 pKa = 6.73GDD841 pKa = 3.43ANYY844 pKa = 10.99LSLATYY850 pKa = 10.56NLEE853 pKa = 4.22NLDD856 pKa = 4.69ASDD859 pKa = 3.53QKK861 pKa = 11.32FDD863 pKa = 3.71ILAADD868 pKa = 3.48IVYY871 pKa = 10.61NLRR874 pKa = 11.84APDD877 pKa = 4.15IIAVQEE883 pKa = 3.97IQDD886 pKa = 3.6ADD888 pKa = 3.94GAGSGSNLSGTVTAQGLIDD907 pKa = 5.58AIYY910 pKa = 9.35AQSGLHH916 pKa = 5.08YY917 pKa = 9.92AYY919 pKa = 9.82IEE921 pKa = 4.15IAPTTAGSTGGEE933 pKa = 3.67PGGNIRR939 pKa = 11.84NGYY942 pKa = 9.03FYY944 pKa = 10.92NIDD947 pKa = 3.16RR948 pKa = 11.84VSYY951 pKa = 11.01VEE953 pKa = 5.82GSAQLIDD960 pKa = 3.28GAAYY964 pKa = 10.35NGTRR968 pKa = 11.84KK969 pKa = 9.67PLVAQFAFAGQTITTIDD986 pKa = 3.21VHH988 pKa = 5.32LTSRR992 pKa = 11.84LGSDD996 pKa = 4.19PLWGDD1001 pKa = 3.35NQPANDD1007 pKa = 4.34AGDD1010 pKa = 3.79AARR1013 pKa = 11.84TAQAAGVKK1021 pKa = 10.03AWVQDD1026 pKa = 3.73HH1027 pKa = 6.65LADD1030 pKa = 4.35NPALNIAVLGDD1041 pKa = 3.21WNGFYY1046 pKa = 10.63FEE1048 pKa = 4.81HH1049 pKa = 7.05AQTQLTDD1056 pKa = 3.64PAQGGVFTNLNTLLPEE1072 pKa = 3.99QEE1074 pKa = 4.27RR1075 pKa = 11.84YY1076 pKa = 10.64SYY1078 pKa = 10.2MFEE1081 pKa = 4.59GNAQQIDD1088 pKa = 4.27NILVTGGLVTNAQYY1102 pKa = 11.32DD1103 pKa = 4.06AVHH1106 pKa = 6.61LNSQFGGSRR1115 pKa = 11.84ATDD1118 pKa = 3.46HH1119 pKa = 7.24DD1120 pKa = 4.35PQVSLLFLGAAPKK1133 pKa = 10.57DD1134 pKa = 3.59LALSNASVAEE1144 pKa = 3.89NLPAGSVVGTVSATDD1159 pKa = 3.45TANDD1163 pKa = 3.8TLHH1166 pKa = 6.48YY1167 pKa = 10.87ALVDD1171 pKa = 3.46NAGGLFAIDD1180 pKa = 3.62AATGVITTTAPFNHH1194 pKa = 6.59EE1195 pKa = 4.0ALAEE1199 pKa = 3.98YY1200 pKa = 11.02ALIAKK1205 pKa = 8.21ATDD1208 pKa = 3.22SGGLTTQQSFTVAVTDD1224 pKa = 3.74VNEE1227 pKa = 4.46APVAAHH1233 pKa = 6.95DD1234 pKa = 3.76AVAVNEE1240 pKa = 4.42DD1241 pKa = 3.41ATTANLWTTLLGNDD1255 pKa = 3.82SDD1257 pKa = 4.73PDD1259 pKa = 3.67VGQTLAISAVDD1270 pKa = 3.32ATGTLGSLVFDD1281 pKa = 4.67AASHH1285 pKa = 4.83TLKK1288 pKa = 10.89YY1289 pKa = 10.59VADD1292 pKa = 4.12ADD1294 pKa = 4.49AFDD1297 pKa = 4.29SLAPGATQVDD1307 pKa = 4.09HH1308 pKa = 7.21FSYY1311 pKa = 10.4TVTDD1315 pKa = 4.17GNGLASTATVDD1326 pKa = 3.52VTVTGIADD1334 pKa = 5.29GITRR1338 pKa = 11.84NGTIFSDD1345 pKa = 3.9TLNGSAGEE1353 pKa = 4.28DD1354 pKa = 3.32RR1355 pKa = 11.84LSGGIGSDD1363 pKa = 3.1TLYY1366 pKa = 11.44GLGGHH1371 pKa = 6.96DD1372 pKa = 4.0WLNGGIGNDD1381 pKa = 3.52KK1382 pKa = 10.96LYY1384 pKa = 11.17GGDD1387 pKa = 3.74GNDD1390 pKa = 3.7VLFGDD1395 pKa = 5.33LGNDD1399 pKa = 3.85TLWGGNGRR1407 pKa = 11.84DD1408 pKa = 3.61VLFGGLGNDD1417 pKa = 3.67TLYY1420 pKa = 11.35GGADD1424 pKa = 3.2ADD1426 pKa = 3.89SFHH1429 pKa = 7.25FGRR1432 pKa = 11.84LEE1434 pKa = 4.12GSATIADD1441 pKa = 3.81FNIAEE1446 pKa = 4.48DD1447 pKa = 5.23KK1448 pKa = 10.83IVLDD1452 pKa = 4.71DD1453 pKa = 4.22GVSVTRR1459 pKa = 11.84TKK1461 pKa = 10.81VQDD1464 pKa = 3.43VNRR1467 pKa = 11.84DD1468 pKa = 3.31GVKK1471 pKa = 10.66DD1472 pKa = 3.62LTLTLSWGSSVTLLGVSDD1490 pKa = 4.33AAQVKK1495 pKa = 10.03YY1496 pKa = 10.27GAPDD1500 pKa = 3.39HH1501 pKa = 6.92YY1502 pKa = 11.02SDD1504 pKa = 4.36HH1505 pKa = 6.37QPGLGGFLDD1514 pKa = 5.42DD1515 pKa = 5.61IGDD1518 pKa = 4.25LFDD1521 pKa = 4.1SLYY1524 pKa = 11.08AGHH1527 pKa = 6.47QKK1529 pKa = 10.9LIDD1532 pKa = 3.74TGWFF1536 pKa = 3.29
MM1 pKa = 7.75PAPAFINEE9 pKa = 3.72IHH11 pKa = 6.67YY12 pKa = 11.02DD13 pKa = 3.48NAGADD18 pKa = 3.16SGEE21 pKa = 4.29FIEE24 pKa = 4.58IAGIAGTDD32 pKa = 3.36LTGWKK37 pKa = 9.47IVLYY41 pKa = 10.42NGANGASYY49 pKa = 10.92NPVMTLSGVIANQQNGFGTISVAYY73 pKa = 7.04PTDD76 pKa = 5.04GIQNGSPDD84 pKa = 3.6AVALVDD90 pKa = 3.72ASNNVVQFLSYY101 pKa = 10.23EE102 pKa = 4.25GSFTATNGPAAGLTSVNVGVSEE124 pKa = 4.59NGAQSGTSIGLVGSGATYY142 pKa = 11.04EE143 pKa = 4.1DD144 pKa = 4.15FHH146 pKa = 6.57WALINGSTAGGVNAGQSFNGVVPPQPGTLSIADD179 pKa = 3.55ATTVEE184 pKa = 4.95GNSGTHH190 pKa = 6.12EE191 pKa = 3.9IVFTVTRR198 pKa = 11.84ADD200 pKa = 3.54GSAGAVSATWTVAFGSADD218 pKa = 3.41AADD221 pKa = 5.0FGAGFTATGTVSFADD236 pKa = 3.79GATTAEE242 pKa = 3.84IRR244 pKa = 11.84LPVQGDD250 pKa = 3.56TAFEE254 pKa = 4.53SNDD257 pKa = 2.94GFTVRR262 pKa = 11.84LSAPQGGVALGDD274 pKa = 3.46AVAAGTITNDD284 pKa = 3.16AAAPPAPPANVFINEE299 pKa = 3.67IHH301 pKa = 6.64YY302 pKa = 11.22DD303 pKa = 3.62NAGTDD308 pKa = 3.05AGEE311 pKa = 4.54AIEE314 pKa = 4.2IAGAAGTDD322 pKa = 3.33LTGYY326 pKa = 10.95KK327 pKa = 10.2LVFYY331 pKa = 10.45NGSNTPGAAPVYY343 pKa = 8.55DD344 pKa = 3.88TLALSGVIDD353 pKa = 4.37DD354 pKa = 4.55EE355 pKa = 5.44SNGFGALGFLRR366 pKa = 11.84AGIQNGAADD375 pKa = 4.78GVALIAPDD383 pKa = 3.82GSVVQLLSYY392 pKa = 10.22EE393 pKa = 4.46GSFTAAAGTPAAGTTSTDD411 pKa = 2.58IGVTEE416 pKa = 4.77EE417 pKa = 3.97PAPAAGLSLQLKK429 pKa = 10.11GSGSSAADD437 pKa = 3.63FAWADD442 pKa = 3.4ASDD445 pKa = 4.28DD446 pKa = 4.11SFGSLNAGQSFLSATGTGHH465 pKa = 7.31LRR467 pKa = 11.84IGDD470 pKa = 3.69ARR472 pKa = 11.84VVEE475 pKa = 4.64GDD477 pKa = 3.25SGTTNLVFTVNRR489 pKa = 11.84AGGTALAATVDD500 pKa = 3.81YY501 pKa = 10.91AVNLDD506 pKa = 3.74GTATLADD513 pKa = 4.51LAPGAVLSGTLSFAPGEE530 pKa = 3.96FSKK533 pKa = 11.09QIVVAVNGDD542 pKa = 3.77LVGEE546 pKa = 4.18PNEE549 pKa = 4.18TLSVSLGATTGNVVIDD565 pKa = 4.1DD566 pKa = 4.05AAATGTITNDD576 pKa = 3.17DD577 pKa = 4.98PIALTISQIQGAGHH591 pKa = 5.89VSAYY595 pKa = 10.14AGQVVITTGIVTAVDD610 pKa = 3.38TNGFYY615 pKa = 10.84LQSAVGDD622 pKa = 3.77GDD624 pKa = 4.29AATSDD629 pKa = 3.29AVFVFTSTAPGVVMGDD645 pKa = 3.58GVSVRR650 pKa = 11.84GSVAEE655 pKa = 4.23FQGSTASLSLTEE667 pKa = 4.35IVAPTVTVEE676 pKa = 3.88THH678 pKa = 6.27GNALPTALLIGTGGILPPSEE698 pKa = 5.17VIDD701 pKa = 4.65DD702 pKa = 5.03DD703 pKa = 5.02GLTSYY708 pKa = 11.34DD709 pKa = 4.01PLHH712 pKa = 7.16DD713 pKa = 5.28GVDD716 pKa = 3.99FWEE719 pKa = 4.5SLEE722 pKa = 3.93GMRR725 pKa = 11.84VTLDD729 pKa = 3.19TPQAVSNTTSFGEE742 pKa = 3.97TDD744 pKa = 3.56VVVSHH749 pKa = 6.65GNGASGINDD758 pKa = 3.41RR759 pKa = 11.84GGITISGDD767 pKa = 3.09GSGVPDD773 pKa = 3.85YY774 pKa = 11.02NPEE777 pKa = 4.5KK778 pKa = 10.6IQIDD782 pKa = 3.88DD783 pKa = 3.97DD784 pKa = 4.42SGIFAGFTPGYY795 pKa = 9.03TIGDD799 pKa = 3.71QLSSVTGVVNYY810 pKa = 10.68AFDD813 pKa = 3.98AYY815 pKa = 10.06EE816 pKa = 4.08VLVTEE821 pKa = 5.03AVTVTQDD828 pKa = 2.95TTLTRR833 pKa = 11.84EE834 pKa = 3.85QTALHH839 pKa = 6.73GDD841 pKa = 3.43ANYY844 pKa = 10.99LSLATYY850 pKa = 10.56NLEE853 pKa = 4.22NLDD856 pKa = 4.69ASDD859 pKa = 3.53QKK861 pKa = 11.32FDD863 pKa = 3.71ILAADD868 pKa = 3.48IVYY871 pKa = 10.61NLRR874 pKa = 11.84APDD877 pKa = 4.15IIAVQEE883 pKa = 3.97IQDD886 pKa = 3.6ADD888 pKa = 3.94GAGSGSNLSGTVTAQGLIDD907 pKa = 5.58AIYY910 pKa = 9.35AQSGLHH916 pKa = 5.08YY917 pKa = 9.92AYY919 pKa = 9.82IEE921 pKa = 4.15IAPTTAGSTGGEE933 pKa = 3.67PGGNIRR939 pKa = 11.84NGYY942 pKa = 9.03FYY944 pKa = 10.92NIDD947 pKa = 3.16RR948 pKa = 11.84VSYY951 pKa = 11.01VEE953 pKa = 5.82GSAQLIDD960 pKa = 3.28GAAYY964 pKa = 10.35NGTRR968 pKa = 11.84KK969 pKa = 9.67PLVAQFAFAGQTITTIDD986 pKa = 3.21VHH988 pKa = 5.32LTSRR992 pKa = 11.84LGSDD996 pKa = 4.19PLWGDD1001 pKa = 3.35NQPANDD1007 pKa = 4.34AGDD1010 pKa = 3.79AARR1013 pKa = 11.84TAQAAGVKK1021 pKa = 10.03AWVQDD1026 pKa = 3.73HH1027 pKa = 6.65LADD1030 pKa = 4.35NPALNIAVLGDD1041 pKa = 3.21WNGFYY1046 pKa = 10.63FEE1048 pKa = 4.81HH1049 pKa = 7.05AQTQLTDD1056 pKa = 3.64PAQGGVFTNLNTLLPEE1072 pKa = 3.99QEE1074 pKa = 4.27RR1075 pKa = 11.84YY1076 pKa = 10.64SYY1078 pKa = 10.2MFEE1081 pKa = 4.59GNAQQIDD1088 pKa = 4.27NILVTGGLVTNAQYY1102 pKa = 11.32DD1103 pKa = 4.06AVHH1106 pKa = 6.61LNSQFGGSRR1115 pKa = 11.84ATDD1118 pKa = 3.46HH1119 pKa = 7.24DD1120 pKa = 4.35PQVSLLFLGAAPKK1133 pKa = 10.57DD1134 pKa = 3.59LALSNASVAEE1144 pKa = 3.89NLPAGSVVGTVSATDD1159 pKa = 3.45TANDD1163 pKa = 3.8TLHH1166 pKa = 6.48YY1167 pKa = 10.87ALVDD1171 pKa = 3.46NAGGLFAIDD1180 pKa = 3.62AATGVITTTAPFNHH1194 pKa = 6.59EE1195 pKa = 4.0ALAEE1199 pKa = 3.98YY1200 pKa = 11.02ALIAKK1205 pKa = 8.21ATDD1208 pKa = 3.22SGGLTTQQSFTVAVTDD1224 pKa = 3.74VNEE1227 pKa = 4.46APVAAHH1233 pKa = 6.95DD1234 pKa = 3.76AVAVNEE1240 pKa = 4.42DD1241 pKa = 3.41ATTANLWTTLLGNDD1255 pKa = 3.82SDD1257 pKa = 4.73PDD1259 pKa = 3.67VGQTLAISAVDD1270 pKa = 3.32ATGTLGSLVFDD1281 pKa = 4.67AASHH1285 pKa = 4.83TLKK1288 pKa = 10.89YY1289 pKa = 10.59VADD1292 pKa = 4.12ADD1294 pKa = 4.49AFDD1297 pKa = 4.29SLAPGATQVDD1307 pKa = 4.09HH1308 pKa = 7.21FSYY1311 pKa = 10.4TVTDD1315 pKa = 4.17GNGLASTATVDD1326 pKa = 3.52VTVTGIADD1334 pKa = 5.29GITRR1338 pKa = 11.84NGTIFSDD1345 pKa = 3.9TLNGSAGEE1353 pKa = 4.28DD1354 pKa = 3.32RR1355 pKa = 11.84LSGGIGSDD1363 pKa = 3.1TLYY1366 pKa = 11.44GLGGHH1371 pKa = 6.96DD1372 pKa = 4.0WLNGGIGNDD1381 pKa = 3.52KK1382 pKa = 10.96LYY1384 pKa = 11.17GGDD1387 pKa = 3.74GNDD1390 pKa = 3.7VLFGDD1395 pKa = 5.33LGNDD1399 pKa = 3.85TLWGGNGRR1407 pKa = 11.84DD1408 pKa = 3.61VLFGGLGNDD1417 pKa = 3.67TLYY1420 pKa = 11.35GGADD1424 pKa = 3.2ADD1426 pKa = 3.89SFHH1429 pKa = 7.25FGRR1432 pKa = 11.84LEE1434 pKa = 4.12GSATIADD1441 pKa = 3.81FNIAEE1446 pKa = 4.48DD1447 pKa = 5.23KK1448 pKa = 10.83IVLDD1452 pKa = 4.71DD1453 pKa = 4.22GVSVTRR1459 pKa = 11.84TKK1461 pKa = 10.81VQDD1464 pKa = 3.43VNRR1467 pKa = 11.84DD1468 pKa = 3.31GVKK1471 pKa = 10.66DD1472 pKa = 3.62LTLTLSWGSSVTLLGVSDD1490 pKa = 4.33AAQVKK1495 pKa = 10.03YY1496 pKa = 10.27GAPDD1500 pKa = 3.39HH1501 pKa = 6.92YY1502 pKa = 11.02SDD1504 pKa = 4.36HH1505 pKa = 6.37QPGLGGFLDD1514 pKa = 5.42DD1515 pKa = 5.61IGDD1518 pKa = 4.25LFDD1521 pKa = 4.1SLYY1524 pKa = 11.08AGHH1527 pKa = 6.47QKK1529 pKa = 10.9LIDD1532 pKa = 3.74TGWFF1536 pKa = 3.29
Molecular weight: 155.92 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q8XRY6|A0A0Q8XRY6_9SPHN Exopolyphosphatase OS=Sphingomonas sp. Root241 OX=1736501 GN=ASE13_00070 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.58GFRR19 pKa = 11.84SRR21 pKa = 11.84MATVGGRR28 pKa = 11.84NVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.58GFRR19 pKa = 11.84SRR21 pKa = 11.84MATVGGRR28 pKa = 11.84NVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
Molecular weight: 5.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1245473 |
41 |
5545 |
335.3 |
36.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.37 ± 0.068 | 0.67 ± 0.013 |
5.783 ± 0.03 | 5.388 ± 0.054 |
3.627 ± 0.027 | 9.239 ± 0.085 |
1.862 ± 0.022 | 4.836 ± 0.027 |
2.907 ± 0.038 | 9.875 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.216 ± 0.023 | 2.765 ± 0.052 |
5.399 ± 0.045 | 3.083 ± 0.026 |
7.261 ± 0.053 | 5.21 ± 0.041 |
5.559 ± 0.068 | 7.199 ± 0.031 |
1.488 ± 0.016 | 2.263 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
