
Ectocarpus siliculosus (Brown alga) (Conferva siliculosa)
Taxonomy: cellular organisms; Eukaryota; Sar; Stramenopiles; Ochrophyta; PX clade; Phaeophyceae; Ectocarpales; Ectocarpaceae; Ectocarpus
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
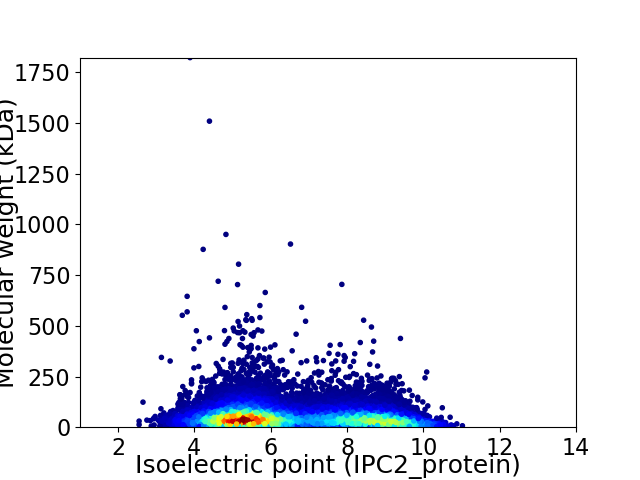
Virtual 2D-PAGE plot for 16334 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D8LQK9|D8LQK9_ECTSI Uncharacterized protein OS=Ectocarpus siliculosus OX=2880 GN=Esi_0006_0175 PE=4 SV=1
MM1 pKa = 7.32EE2 pKa = 4.66VVLSPEE8 pKa = 4.18DD9 pKa = 3.58LEE11 pKa = 5.78GYY13 pKa = 8.32PSPGDD18 pKa = 3.33AVVYY22 pKa = 8.92TITITNDD29 pKa = 2.92GSLTLHH35 pKa = 7.04DD36 pKa = 4.45VTPDD40 pKa = 3.34SPQALAWACEE50 pKa = 4.0LAEE53 pKa = 4.51GAALVPGVEE62 pKa = 4.61TKK64 pKa = 10.8CVGRR68 pKa = 11.84LVLTQNNLDD77 pKa = 3.71EE78 pKa = 4.86AQVVTTVTITATDD91 pKa = 3.51SANEE95 pKa = 3.95LVEE98 pKa = 6.09DD99 pKa = 4.6DD100 pKa = 4.55LSSTVEE106 pKa = 4.27LPTSSSLLVVEE117 pKa = 4.94TCSFTGSDD125 pKa = 3.26ANRR128 pKa = 11.84AEE130 pKa = 4.27VGQGVAYY137 pKa = 8.84TFTMKK142 pKa = 9.58NTGSTTLTGLDD153 pKa = 3.71VNSVFLDD160 pKa = 3.48QALDD164 pKa = 3.72NVACGASSLSEE175 pKa = 4.03LARR178 pKa = 11.84DD179 pKa = 3.77DD180 pKa = 4.7TIVCSSVLDD189 pKa = 3.78HH190 pKa = 7.72AITQVSAVASDD201 pKa = 3.25STMVSEE207 pKa = 4.78GVSISTCVDD216 pKa = 4.47GISSLSLGKK225 pKa = 8.45TVVGGGSWVDD235 pKa = 3.53SNGDD239 pKa = 3.37EE240 pKa = 4.2DD241 pKa = 4.11TDD243 pKa = 3.7AGEE246 pKa = 4.3VILYY250 pKa = 9.47QLLVVNVGTVTLNSVVLTDD269 pKa = 4.32GSVTSEE275 pKa = 4.04GVSCEE280 pKa = 3.86SGIPDD285 pKa = 3.62SLLPGEE291 pKa = 4.85GFEE294 pKa = 4.65CQAEE298 pKa = 4.16YY299 pKa = 9.97TIVQDD304 pKa = 3.64DD305 pKa = 3.79VDD307 pKa = 4.01RR308 pKa = 11.84GFVVSNATVTAINPSKK324 pKa = 11.09DD325 pKa = 3.28KK326 pKa = 10.75TNKK329 pKa = 8.34TSEE332 pKa = 3.95VSTDD336 pKa = 3.38LVRR339 pKa = 11.84RR340 pKa = 11.84PAISLDD346 pKa = 4.02TVGSWANAGNGSDD359 pKa = 3.74GFADD363 pKa = 3.81AGDD366 pKa = 3.79TAVYY370 pKa = 9.81RR371 pKa = 11.84YY372 pKa = 7.8TSSEE376 pKa = 3.99PGQSAGCSGLSVLNWAAIEE395 pKa = 4.26AGGLTTVSRR404 pKa = 11.84VHH406 pKa = 6.96SVDD409 pKa = 2.96ATLGLPVPSQTTVSIDD425 pKa = 3.72LPPPPSIQLDD435 pKa = 3.83MVGTFADD442 pKa = 4.41DD443 pKa = 3.98SADD446 pKa = 3.57GMQGLADD453 pKa = 3.8VGEE456 pKa = 4.38MISYY460 pKa = 10.15VFTITNNGHH469 pKa = 6.47AVLEE473 pKa = 5.15GITLADD479 pKa = 3.72GGTNAGISSTTTCGDD494 pKa = 3.22TAIASRR500 pKa = 11.84NASSITVGGTLAVRR514 pKa = 11.84AVITCNSSYY523 pKa = 11.11AITEE527 pKa = 4.31DD528 pKa = 3.3NVNALEE534 pKa = 4.18VSSAASVTASDD545 pKa = 3.81VSGNAVDD552 pKa = 4.77AADD555 pKa = 3.75ATVVSLDD562 pKa = 4.08QINTGTKK569 pKa = 9.49SAGAQVTAWYY579 pKa = 9.77EE580 pKa = 3.95DD581 pKa = 3.77EE582 pKa = 5.02PGNLSEE588 pKa = 4.35EE589 pKa = 4.45TSDD592 pKa = 3.53GTVVDD597 pKa = 4.21VALKK601 pKa = 9.93QDD603 pKa = 3.89PLAGITKK610 pKa = 8.67TFVFTPARR618 pKa = 11.84EE619 pKa = 4.08DD620 pKa = 3.32GMASVDD626 pKa = 3.39DD627 pKa = 4.77TIAYY631 pKa = 7.74TITASNDD638 pKa = 3.26GNVDD642 pKa = 3.45LSDD645 pKa = 3.11ITVTDD650 pKa = 3.56EE651 pKa = 5.52RR652 pKa = 11.84FLNPQGGFDD661 pKa = 4.39MNWEE665 pKa = 4.22SDD667 pKa = 3.42TSFGMLPGAEE677 pKa = 4.01LHH679 pKa = 6.86CYY681 pKa = 6.99PTTAITQADD690 pKa = 3.23IDD692 pKa = 4.2AGVVASNVTHH702 pKa = 7.28PDD704 pKa = 3.0AAIVCTPSLLGLSLAPGAEE723 pKa = 4.5VSCSAFYY730 pKa = 10.03PVSQDD735 pKa = 3.2DD736 pKa = 3.83VNAGVVSSEE745 pKa = 3.89ATISADD751 pKa = 3.66SPIGPISVSNSSEE764 pKa = 4.06PQSLEE769 pKa = 3.72QVDD772 pKa = 5.01GIDD775 pKa = 3.76IGDD778 pKa = 3.94EE779 pKa = 4.2LFCPLEE785 pKa = 4.08GHH787 pKa = 6.41TFTCTAVVHH796 pKa = 5.32VTQEE800 pKa = 3.87NMNDD804 pKa = 3.05GHH806 pKa = 6.99VDD808 pKa = 3.01HH809 pKa = 7.93DD810 pKa = 3.74IGVTAKK816 pKa = 9.6TAIGDD821 pKa = 3.86EE822 pKa = 4.35PLDD825 pKa = 3.71DD826 pKa = 3.9QYY828 pKa = 11.95RR829 pKa = 11.84LHH831 pKa = 6.7VPLEE835 pKa = 4.37GKK837 pKa = 9.55SAFLIEE843 pKa = 4.84HH844 pKa = 6.12NSEE847 pKa = 4.07YY848 pKa = 10.99LPVDD852 pKa = 4.46GIGAASALGDD862 pKa = 4.09EE863 pKa = 4.57ITYY866 pKa = 8.86TLNIDD871 pKa = 3.56NNGTVTLSSVTPVNSKK887 pKa = 10.47VEE889 pKa = 4.21LTCEE893 pKa = 3.9PDD895 pKa = 3.53VNSGAKK901 pKa = 9.86LDD903 pKa = 4.03AGEE906 pKa = 4.65GAMCTGTYY914 pKa = 9.94IVTQDD919 pKa = 4.83DD920 pKa = 3.31IDD922 pKa = 3.86AGKK925 pKa = 9.64IVCAASVTATDD936 pKa = 4.33PDD938 pKa = 3.93GEE940 pKa = 4.84LIFRR944 pKa = 11.84QTRR947 pKa = 11.84ISQHH951 pKa = 5.0VSQTPQLSVVLSSVHH966 pKa = 5.4TVNSLDD972 pKa = 3.25GKK974 pKa = 6.96TRR976 pKa = 11.84KK977 pKa = 9.99GDD979 pKa = 3.44TVLYY983 pKa = 5.85TTQVFNSGNTCLTDD997 pKa = 3.48VKK999 pKa = 10.52ISEE1002 pKa = 4.42LLVDD1006 pKa = 4.78GGLDD1010 pKa = 3.53CGSASSTLCPTDD1022 pKa = 3.41EE1023 pKa = 5.77AISCTGTHH1031 pKa = 6.62TLTQANVDD1039 pKa = 3.77SVHH1042 pKa = 7.24IINTATATASPLFANTSDD1060 pKa = 3.65EE1061 pKa = 4.72SNMISAGDD1069 pKa = 3.8GDD1071 pKa = 4.4TVSWLLYY1078 pKa = 8.75PAISVGSLTSVSLEE1092 pKa = 3.83VLLLEE1097 pKa = 4.46RR1098 pKa = 11.84SGTIVDD1104 pKa = 4.07CTPSVTEE1111 pKa = 4.54ALVLAPGGIVLCSATLEE1128 pKa = 4.19LTQDD1132 pKa = 4.19HH1133 pKa = 7.25IDD1135 pKa = 3.85GGALTSEE1142 pKa = 4.07MFARR1146 pKa = 11.84GVASDD1151 pKa = 3.7GQAVLGEE1158 pKa = 4.3DD1159 pKa = 3.5SVHH1162 pKa = 6.22QEE1164 pKa = 3.8LVQDD1168 pKa = 3.78VGLSVAEE1175 pKa = 3.69IGAFNDD1181 pKa = 3.16EE1182 pKa = 5.17DD1183 pKa = 4.75GDD1185 pKa = 4.02EE1186 pKa = 4.81LGDD1189 pKa = 3.81AGEE1192 pKa = 4.4TMFYY1196 pKa = 10.13TATFRR1201 pKa = 11.84NIGNVRR1207 pKa = 11.84VGNVRR1212 pKa = 11.84VSHH1215 pKa = 5.69LQGHH1219 pKa = 6.52IAALACDD1226 pKa = 3.72SGFEE1230 pKa = 4.54AVTSTDD1236 pKa = 3.0GLGEE1240 pKa = 4.3LLPGIEE1246 pKa = 4.33FGCRR1250 pKa = 11.84ATYY1253 pKa = 10.92SLTQADD1259 pKa = 3.22VDD1261 pKa = 3.65AAEE1264 pKa = 4.46VVNTASISGVARR1276 pKa = 11.84DD1277 pKa = 3.69TSGTEE1282 pKa = 3.91VEE1284 pKa = 6.01AEE1286 pKa = 4.11DD1287 pKa = 3.64SWKK1290 pKa = 10.35QEE1292 pKa = 3.88YY1293 pKa = 7.99TQLAMALLNVVGVFHH1308 pKa = 7.9DD1309 pKa = 3.69VGTVDD1314 pKa = 5.79DD1315 pKa = 4.79EE1316 pKa = 5.69ADD1318 pKa = 2.98IHH1320 pKa = 7.29DD1321 pKa = 3.78KK1322 pKa = 10.7MEE1324 pKa = 4.19YY1325 pKa = 8.96TVEE1328 pKa = 3.85IANTGTVTLTDD1339 pKa = 3.44VGVTGSLFEE1348 pKa = 4.74NEE1350 pKa = 5.34AIPCPGATLAPAEE1363 pKa = 4.39SMVCNASYY1371 pKa = 10.12TITNTDD1377 pKa = 2.55IDD1379 pKa = 4.13RR1380 pKa = 11.84YY1381 pKa = 9.29EE1382 pKa = 4.23VEE1384 pKa = 4.09TTADD1388 pKa = 3.52VTASGPFAQAVGDD1401 pKa = 3.82SGVYY1405 pKa = 9.7LQRR1408 pKa = 11.84LDD1410 pKa = 3.92AQPSVSLLMEE1420 pKa = 4.64GVHH1423 pKa = 6.51VDD1425 pKa = 3.46SQIDD1429 pKa = 3.63STASAEE1435 pKa = 4.13SDD1437 pKa = 3.61GNSVADD1443 pKa = 3.57AGEE1446 pKa = 4.06YY1447 pKa = 8.83TRR1449 pKa = 11.84YY1450 pKa = 8.75TMVVRR1455 pKa = 11.84NTGMLTVHH1463 pKa = 6.72QITVVEE1469 pKa = 4.17SLEE1472 pKa = 4.38GGEE1475 pKa = 4.1ATCPKK1480 pKa = 10.35NYY1482 pKa = 10.75LEE1484 pKa = 4.33MAEE1487 pKa = 4.69SMVCNATYY1495 pKa = 10.49PLTQEE1500 pKa = 4.31DD1501 pKa = 4.11VDD1503 pKa = 4.48RR1504 pKa = 11.84GDD1506 pKa = 3.54VTSVTTLDD1514 pKa = 3.34AFGPTRR1520 pKa = 11.84DD1521 pKa = 3.92DD1522 pKa = 3.63GEE1524 pKa = 4.3ARR1526 pKa = 11.84STTRR1530 pKa = 11.84AEE1532 pKa = 3.69ASSWVKK1538 pKa = 10.58LPQDD1542 pKa = 3.43PSILVTKK1549 pKa = 9.74EE1550 pKa = 3.87CAWQDD1555 pKa = 3.12GAEE1558 pKa = 4.16RR1559 pKa = 11.84DD1560 pKa = 3.96GLPDD1564 pKa = 4.16PGEE1567 pKa = 4.27VVVLIYY1573 pKa = 9.95TVSNTGSVTLTDD1585 pKa = 4.14GSLQQSTDD1593 pKa = 2.99LGLQVVDD1600 pKa = 4.8CMNAIPSAGNVVCTSSIEE1618 pKa = 3.88IGQADD1623 pKa = 3.82IDD1625 pKa = 4.24AGTVTSTVEE1634 pKa = 3.69ITALSPLGDD1643 pKa = 3.65MTGSTTNCSWAWGVLPAQVDD1663 pKa = 3.87TVITGRR1669 pKa = 11.84FVDD1672 pKa = 3.9GDD1674 pKa = 3.7GDD1676 pKa = 4.36GIASVGEE1683 pKa = 3.94GTGYY1687 pKa = 11.04NVTIVNNGKK1696 pKa = 8.35VTVTVVEE1703 pKa = 4.22VDD1705 pKa = 3.22ATAVARR1711 pKa = 11.84GSPSSGNSASGVTIVCEE1728 pKa = 4.07PLLDD1732 pKa = 4.01TSPLLAPGQSLEE1744 pKa = 4.28CTAEE1748 pKa = 3.66IDD1750 pKa = 4.49IDD1752 pKa = 3.73QDD1754 pKa = 3.56DD1755 pKa = 4.14VNRR1758 pKa = 11.84GEE1760 pKa = 4.22IEE1762 pKa = 4.26LEE1764 pKa = 4.09FEE1766 pKa = 4.35VTADD1770 pKa = 3.55NPAGDD1775 pKa = 4.28EE1776 pKa = 4.33STVTDD1781 pKa = 3.65SSSIEE1786 pKa = 4.0LPAKK1790 pKa = 10.29YY1791 pKa = 9.95RR1792 pKa = 11.84IALSLEE1798 pKa = 4.15GSNTTSSDD1806 pKa = 3.25RR1807 pKa = 11.84DD1808 pKa = 3.64DD1809 pKa = 4.82DD1810 pKa = 4.58GKK1812 pKa = 11.3SSSGDD1817 pKa = 3.16EE1818 pKa = 4.05VTFTLTVANRR1828 pKa = 11.84GSVDD1832 pKa = 3.51LTGVEE1837 pKa = 3.99AQVAGLEE1844 pKa = 4.22GLVCQQFIPSTVGQRR1859 pKa = 11.84RR1860 pKa = 11.84LSWAEE1865 pKa = 3.81TNTDD1869 pKa = 2.91QDD1871 pKa = 3.23IFPPDD1876 pKa = 3.31EE1877 pKa = 5.03EE1878 pKa = 4.78YY1879 pKa = 10.59TCTASHH1885 pKa = 7.55ILTQDD1890 pKa = 3.38DD1891 pKa = 3.58VDD1893 pKa = 4.51AGTLYY1898 pKa = 9.37RR1899 pKa = 11.84TASVSSQARR1908 pKa = 11.84DD1909 pKa = 3.41PTGTLVVAGAEE1920 pKa = 4.06EE1921 pKa = 4.2TLALVAYY1928 pKa = 7.25PAIVVVIVSGHH1939 pKa = 4.72AA1940 pKa = 3.0
MM1 pKa = 7.32EE2 pKa = 4.66VVLSPEE8 pKa = 4.18DD9 pKa = 3.58LEE11 pKa = 5.78GYY13 pKa = 8.32PSPGDD18 pKa = 3.33AVVYY22 pKa = 8.92TITITNDD29 pKa = 2.92GSLTLHH35 pKa = 7.04DD36 pKa = 4.45VTPDD40 pKa = 3.34SPQALAWACEE50 pKa = 4.0LAEE53 pKa = 4.51GAALVPGVEE62 pKa = 4.61TKK64 pKa = 10.8CVGRR68 pKa = 11.84LVLTQNNLDD77 pKa = 3.71EE78 pKa = 4.86AQVVTTVTITATDD91 pKa = 3.51SANEE95 pKa = 3.95LVEE98 pKa = 6.09DD99 pKa = 4.6DD100 pKa = 4.55LSSTVEE106 pKa = 4.27LPTSSSLLVVEE117 pKa = 4.94TCSFTGSDD125 pKa = 3.26ANRR128 pKa = 11.84AEE130 pKa = 4.27VGQGVAYY137 pKa = 8.84TFTMKK142 pKa = 9.58NTGSTTLTGLDD153 pKa = 3.71VNSVFLDD160 pKa = 3.48QALDD164 pKa = 3.72NVACGASSLSEE175 pKa = 4.03LARR178 pKa = 11.84DD179 pKa = 3.77DD180 pKa = 4.7TIVCSSVLDD189 pKa = 3.78HH190 pKa = 7.72AITQVSAVASDD201 pKa = 3.25STMVSEE207 pKa = 4.78GVSISTCVDD216 pKa = 4.47GISSLSLGKK225 pKa = 8.45TVVGGGSWVDD235 pKa = 3.53SNGDD239 pKa = 3.37EE240 pKa = 4.2DD241 pKa = 4.11TDD243 pKa = 3.7AGEE246 pKa = 4.3VILYY250 pKa = 9.47QLLVVNVGTVTLNSVVLTDD269 pKa = 4.32GSVTSEE275 pKa = 4.04GVSCEE280 pKa = 3.86SGIPDD285 pKa = 3.62SLLPGEE291 pKa = 4.85GFEE294 pKa = 4.65CQAEE298 pKa = 4.16YY299 pKa = 9.97TIVQDD304 pKa = 3.64DD305 pKa = 3.79VDD307 pKa = 4.01RR308 pKa = 11.84GFVVSNATVTAINPSKK324 pKa = 11.09DD325 pKa = 3.28KK326 pKa = 10.75TNKK329 pKa = 8.34TSEE332 pKa = 3.95VSTDD336 pKa = 3.38LVRR339 pKa = 11.84RR340 pKa = 11.84PAISLDD346 pKa = 4.02TVGSWANAGNGSDD359 pKa = 3.74GFADD363 pKa = 3.81AGDD366 pKa = 3.79TAVYY370 pKa = 9.81RR371 pKa = 11.84YY372 pKa = 7.8TSSEE376 pKa = 3.99PGQSAGCSGLSVLNWAAIEE395 pKa = 4.26AGGLTTVSRR404 pKa = 11.84VHH406 pKa = 6.96SVDD409 pKa = 2.96ATLGLPVPSQTTVSIDD425 pKa = 3.72LPPPPSIQLDD435 pKa = 3.83MVGTFADD442 pKa = 4.41DD443 pKa = 3.98SADD446 pKa = 3.57GMQGLADD453 pKa = 3.8VGEE456 pKa = 4.38MISYY460 pKa = 10.15VFTITNNGHH469 pKa = 6.47AVLEE473 pKa = 5.15GITLADD479 pKa = 3.72GGTNAGISSTTTCGDD494 pKa = 3.22TAIASRR500 pKa = 11.84NASSITVGGTLAVRR514 pKa = 11.84AVITCNSSYY523 pKa = 11.11AITEE527 pKa = 4.31DD528 pKa = 3.3NVNALEE534 pKa = 4.18VSSAASVTASDD545 pKa = 3.81VSGNAVDD552 pKa = 4.77AADD555 pKa = 3.75ATVVSLDD562 pKa = 4.08QINTGTKK569 pKa = 9.49SAGAQVTAWYY579 pKa = 9.77EE580 pKa = 3.95DD581 pKa = 3.77EE582 pKa = 5.02PGNLSEE588 pKa = 4.35EE589 pKa = 4.45TSDD592 pKa = 3.53GTVVDD597 pKa = 4.21VALKK601 pKa = 9.93QDD603 pKa = 3.89PLAGITKK610 pKa = 8.67TFVFTPARR618 pKa = 11.84EE619 pKa = 4.08DD620 pKa = 3.32GMASVDD626 pKa = 3.39DD627 pKa = 4.77TIAYY631 pKa = 7.74TITASNDD638 pKa = 3.26GNVDD642 pKa = 3.45LSDD645 pKa = 3.11ITVTDD650 pKa = 3.56EE651 pKa = 5.52RR652 pKa = 11.84FLNPQGGFDD661 pKa = 4.39MNWEE665 pKa = 4.22SDD667 pKa = 3.42TSFGMLPGAEE677 pKa = 4.01LHH679 pKa = 6.86CYY681 pKa = 6.99PTTAITQADD690 pKa = 3.23IDD692 pKa = 4.2AGVVASNVTHH702 pKa = 7.28PDD704 pKa = 3.0AAIVCTPSLLGLSLAPGAEE723 pKa = 4.5VSCSAFYY730 pKa = 10.03PVSQDD735 pKa = 3.2DD736 pKa = 3.83VNAGVVSSEE745 pKa = 3.89ATISADD751 pKa = 3.66SPIGPISVSNSSEE764 pKa = 4.06PQSLEE769 pKa = 3.72QVDD772 pKa = 5.01GIDD775 pKa = 3.76IGDD778 pKa = 3.94EE779 pKa = 4.2LFCPLEE785 pKa = 4.08GHH787 pKa = 6.41TFTCTAVVHH796 pKa = 5.32VTQEE800 pKa = 3.87NMNDD804 pKa = 3.05GHH806 pKa = 6.99VDD808 pKa = 3.01HH809 pKa = 7.93DD810 pKa = 3.74IGVTAKK816 pKa = 9.6TAIGDD821 pKa = 3.86EE822 pKa = 4.35PLDD825 pKa = 3.71DD826 pKa = 3.9QYY828 pKa = 11.95RR829 pKa = 11.84LHH831 pKa = 6.7VPLEE835 pKa = 4.37GKK837 pKa = 9.55SAFLIEE843 pKa = 4.84HH844 pKa = 6.12NSEE847 pKa = 4.07YY848 pKa = 10.99LPVDD852 pKa = 4.46GIGAASALGDD862 pKa = 4.09EE863 pKa = 4.57ITYY866 pKa = 8.86TLNIDD871 pKa = 3.56NNGTVTLSSVTPVNSKK887 pKa = 10.47VEE889 pKa = 4.21LTCEE893 pKa = 3.9PDD895 pKa = 3.53VNSGAKK901 pKa = 9.86LDD903 pKa = 4.03AGEE906 pKa = 4.65GAMCTGTYY914 pKa = 9.94IVTQDD919 pKa = 4.83DD920 pKa = 3.31IDD922 pKa = 3.86AGKK925 pKa = 9.64IVCAASVTATDD936 pKa = 4.33PDD938 pKa = 3.93GEE940 pKa = 4.84LIFRR944 pKa = 11.84QTRR947 pKa = 11.84ISQHH951 pKa = 5.0VSQTPQLSVVLSSVHH966 pKa = 5.4TVNSLDD972 pKa = 3.25GKK974 pKa = 6.96TRR976 pKa = 11.84KK977 pKa = 9.99GDD979 pKa = 3.44TVLYY983 pKa = 5.85TTQVFNSGNTCLTDD997 pKa = 3.48VKK999 pKa = 10.52ISEE1002 pKa = 4.42LLVDD1006 pKa = 4.78GGLDD1010 pKa = 3.53CGSASSTLCPTDD1022 pKa = 3.41EE1023 pKa = 5.77AISCTGTHH1031 pKa = 6.62TLTQANVDD1039 pKa = 3.77SVHH1042 pKa = 7.24IINTATATASPLFANTSDD1060 pKa = 3.65EE1061 pKa = 4.72SNMISAGDD1069 pKa = 3.8GDD1071 pKa = 4.4TVSWLLYY1078 pKa = 8.75PAISVGSLTSVSLEE1092 pKa = 3.83VLLLEE1097 pKa = 4.46RR1098 pKa = 11.84SGTIVDD1104 pKa = 4.07CTPSVTEE1111 pKa = 4.54ALVLAPGGIVLCSATLEE1128 pKa = 4.19LTQDD1132 pKa = 4.19HH1133 pKa = 7.25IDD1135 pKa = 3.85GGALTSEE1142 pKa = 4.07MFARR1146 pKa = 11.84GVASDD1151 pKa = 3.7GQAVLGEE1158 pKa = 4.3DD1159 pKa = 3.5SVHH1162 pKa = 6.22QEE1164 pKa = 3.8LVQDD1168 pKa = 3.78VGLSVAEE1175 pKa = 3.69IGAFNDD1181 pKa = 3.16EE1182 pKa = 5.17DD1183 pKa = 4.75GDD1185 pKa = 4.02EE1186 pKa = 4.81LGDD1189 pKa = 3.81AGEE1192 pKa = 4.4TMFYY1196 pKa = 10.13TATFRR1201 pKa = 11.84NIGNVRR1207 pKa = 11.84VGNVRR1212 pKa = 11.84VSHH1215 pKa = 5.69LQGHH1219 pKa = 6.52IAALACDD1226 pKa = 3.72SGFEE1230 pKa = 4.54AVTSTDD1236 pKa = 3.0GLGEE1240 pKa = 4.3LLPGIEE1246 pKa = 4.33FGCRR1250 pKa = 11.84ATYY1253 pKa = 10.92SLTQADD1259 pKa = 3.22VDD1261 pKa = 3.65AAEE1264 pKa = 4.46VVNTASISGVARR1276 pKa = 11.84DD1277 pKa = 3.69TSGTEE1282 pKa = 3.91VEE1284 pKa = 6.01AEE1286 pKa = 4.11DD1287 pKa = 3.64SWKK1290 pKa = 10.35QEE1292 pKa = 3.88YY1293 pKa = 7.99TQLAMALLNVVGVFHH1308 pKa = 7.9DD1309 pKa = 3.69VGTVDD1314 pKa = 5.79DD1315 pKa = 4.79EE1316 pKa = 5.69ADD1318 pKa = 2.98IHH1320 pKa = 7.29DD1321 pKa = 3.78KK1322 pKa = 10.7MEE1324 pKa = 4.19YY1325 pKa = 8.96TVEE1328 pKa = 3.85IANTGTVTLTDD1339 pKa = 3.44VGVTGSLFEE1348 pKa = 4.74NEE1350 pKa = 5.34AIPCPGATLAPAEE1363 pKa = 4.39SMVCNASYY1371 pKa = 10.12TITNTDD1377 pKa = 2.55IDD1379 pKa = 4.13RR1380 pKa = 11.84YY1381 pKa = 9.29EE1382 pKa = 4.23VEE1384 pKa = 4.09TTADD1388 pKa = 3.52VTASGPFAQAVGDD1401 pKa = 3.82SGVYY1405 pKa = 9.7LQRR1408 pKa = 11.84LDD1410 pKa = 3.92AQPSVSLLMEE1420 pKa = 4.64GVHH1423 pKa = 6.51VDD1425 pKa = 3.46SQIDD1429 pKa = 3.63STASAEE1435 pKa = 4.13SDD1437 pKa = 3.61GNSVADD1443 pKa = 3.57AGEE1446 pKa = 4.06YY1447 pKa = 8.83TRR1449 pKa = 11.84YY1450 pKa = 8.75TMVVRR1455 pKa = 11.84NTGMLTVHH1463 pKa = 6.72QITVVEE1469 pKa = 4.17SLEE1472 pKa = 4.38GGEE1475 pKa = 4.1ATCPKK1480 pKa = 10.35NYY1482 pKa = 10.75LEE1484 pKa = 4.33MAEE1487 pKa = 4.69SMVCNATYY1495 pKa = 10.49PLTQEE1500 pKa = 4.31DD1501 pKa = 4.11VDD1503 pKa = 4.48RR1504 pKa = 11.84GDD1506 pKa = 3.54VTSVTTLDD1514 pKa = 3.34AFGPTRR1520 pKa = 11.84DD1521 pKa = 3.92DD1522 pKa = 3.63GEE1524 pKa = 4.3ARR1526 pKa = 11.84STTRR1530 pKa = 11.84AEE1532 pKa = 3.69ASSWVKK1538 pKa = 10.58LPQDD1542 pKa = 3.43PSILVTKK1549 pKa = 9.74EE1550 pKa = 3.87CAWQDD1555 pKa = 3.12GAEE1558 pKa = 4.16RR1559 pKa = 11.84DD1560 pKa = 3.96GLPDD1564 pKa = 4.16PGEE1567 pKa = 4.27VVVLIYY1573 pKa = 9.95TVSNTGSVTLTDD1585 pKa = 4.14GSLQQSTDD1593 pKa = 2.99LGLQVVDD1600 pKa = 4.8CMNAIPSAGNVVCTSSIEE1618 pKa = 3.88IGQADD1623 pKa = 3.82IDD1625 pKa = 4.24AGTVTSTVEE1634 pKa = 3.69ITALSPLGDD1643 pKa = 3.65MTGSTTNCSWAWGVLPAQVDD1663 pKa = 3.87TVITGRR1669 pKa = 11.84FVDD1672 pKa = 3.9GDD1674 pKa = 3.7GDD1676 pKa = 4.36GIASVGEE1683 pKa = 3.94GTGYY1687 pKa = 11.04NVTIVNNGKK1696 pKa = 8.35VTVTVVEE1703 pKa = 4.22VDD1705 pKa = 3.22ATAVARR1711 pKa = 11.84GSPSSGNSASGVTIVCEE1728 pKa = 4.07PLLDD1732 pKa = 4.01TSPLLAPGQSLEE1744 pKa = 4.28CTAEE1748 pKa = 3.66IDD1750 pKa = 4.49IDD1752 pKa = 3.73QDD1754 pKa = 3.56DD1755 pKa = 4.14VNRR1758 pKa = 11.84GEE1760 pKa = 4.22IEE1762 pKa = 4.26LEE1764 pKa = 4.09FEE1766 pKa = 4.35VTADD1770 pKa = 3.55NPAGDD1775 pKa = 4.28EE1776 pKa = 4.33STVTDD1781 pKa = 3.65SSSIEE1786 pKa = 4.0LPAKK1790 pKa = 10.29YY1791 pKa = 9.95RR1792 pKa = 11.84IALSLEE1798 pKa = 4.15GSNTTSSDD1806 pKa = 3.25RR1807 pKa = 11.84DD1808 pKa = 3.64DD1809 pKa = 4.82DD1810 pKa = 4.58GKK1812 pKa = 11.3SSSGDD1817 pKa = 3.16EE1818 pKa = 4.05VTFTLTVANRR1828 pKa = 11.84GSVDD1832 pKa = 3.51LTGVEE1837 pKa = 3.99AQVAGLEE1844 pKa = 4.22GLVCQQFIPSTVGQRR1859 pKa = 11.84RR1860 pKa = 11.84LSWAEE1865 pKa = 3.81TNTDD1869 pKa = 2.91QDD1871 pKa = 3.23IFPPDD1876 pKa = 3.31EE1877 pKa = 5.03EE1878 pKa = 4.78YY1879 pKa = 10.59TCTASHH1885 pKa = 7.55ILTQDD1890 pKa = 3.38DD1891 pKa = 3.58VDD1893 pKa = 4.51AGTLYY1898 pKa = 9.37RR1899 pKa = 11.84TASVSSQARR1908 pKa = 11.84DD1909 pKa = 3.41PTGTLVVAGAEE1920 pKa = 4.06EE1921 pKa = 4.2TLALVAYY1928 pKa = 7.25PAIVVVIVSGHH1939 pKa = 4.72AA1940 pKa = 3.0
Molecular weight: 200.52 kDa
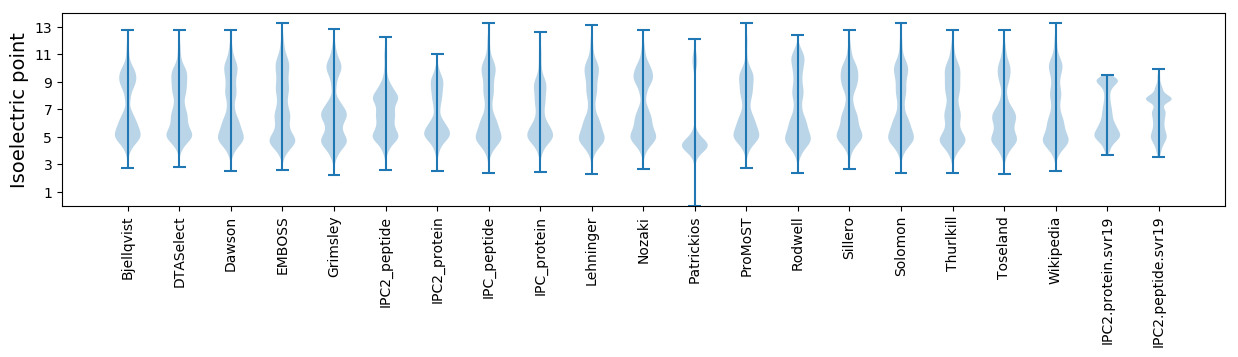
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D8LM76|D8LM76_ECTSI Uncharacterized protein OS=Ectocarpus siliculosus OX=2880 GN=Esi_0392_0004 PE=4 SV=1
MM1 pKa = 7.57AEE3 pKa = 3.94SRR5 pKa = 11.84RR6 pKa = 11.84TIATTTTKK14 pKa = 10.19RR15 pKa = 11.84GTMATSWMTTKK26 pKa = 10.52RR27 pKa = 11.84RR28 pKa = 11.84TTARR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84ATPTRR39 pKa = 11.84TTTTTRR45 pKa = 11.84RR46 pKa = 11.84SRR48 pKa = 11.84SKK50 pKa = 10.97AFRR53 pKa = 11.84TTVSPRR59 pKa = 11.84SKK61 pKa = 10.77SS62 pKa = 3.3
MM1 pKa = 7.57AEE3 pKa = 3.94SRR5 pKa = 11.84RR6 pKa = 11.84TIATTTTKK14 pKa = 10.19RR15 pKa = 11.84GTMATSWMTTKK26 pKa = 10.52RR27 pKa = 11.84RR28 pKa = 11.84TTARR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84ATPTRR39 pKa = 11.84TTTTTRR45 pKa = 11.84RR46 pKa = 11.84SRR48 pKa = 11.84SKK50 pKa = 10.97AFRR53 pKa = 11.84TTVSPRR59 pKa = 11.84SKK61 pKa = 10.77SS62 pKa = 3.3
Molecular weight: 7.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
8478560 |
22 |
17422 |
519.1 |
55.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.427 ± 0.035 | 1.558 ± 0.01 |
5.673 ± 0.019 | 6.842 ± 0.023 |
2.906 ± 0.013 | 10.541 ± 0.036 |
1.98 ± 0.008 | 2.893 ± 0.014 |
4.139 ± 0.021 | 8.126 ± 0.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.131 ± 0.01 | 2.688 ± 0.01 |
5.423 ± 0.025 | 3.29 ± 0.013 |
7.156 ± 0.028 | 7.975 ± 0.024 |
5.315 ± 0.018 | 6.882 ± 0.023 |
1.229 ± 0.007 | 1.821 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
