
Pacific flying fox faeces associated gemycircularvirus-4
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Cyclovirus; unclassified Cyclovirus
Average proteome isoelectric point is 7.09
Get precalculated fractions of proteins
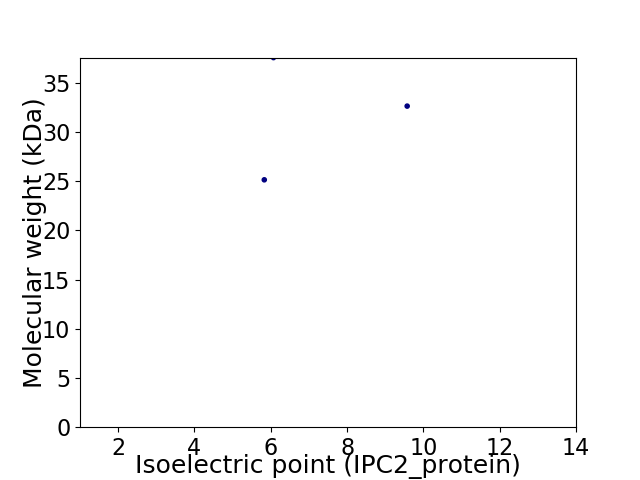
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140CTM2|A0A140CTM2_9CIRC RepA OS=Pacific flying fox faeces associated gemycircularvirus-4 OX=1795996 PE=3 SV=1
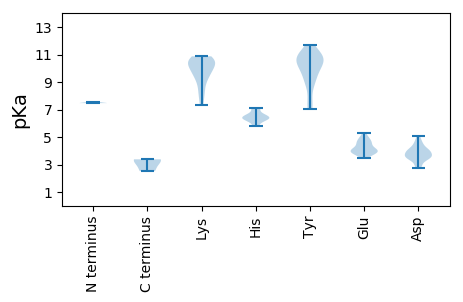
MM1 pKa = 7.47TFLFSARR8 pKa = 11.84YY9 pKa = 9.34VLLTYY14 pKa = 7.24PQCGTLDD21 pKa = 3.23GWAVSDD27 pKa = 4.02HH28 pKa = 6.52LSALGAEE35 pKa = 4.64CIVGRR40 pKa = 11.84EE41 pKa = 4.0NHH43 pKa = 6.3SDD45 pKa = 3.41GGTHH49 pKa = 6.32LHH51 pKa = 6.56AFVDD55 pKa = 4.89FGRR58 pKa = 11.84KK59 pKa = 7.96KK60 pKa = 9.87QSRR63 pKa = 11.84RR64 pKa = 11.84SDD66 pKa = 3.46FFDD69 pKa = 3.73VGGHH73 pKa = 6.35HH74 pKa = 7.14PNIAPSRR81 pKa = 11.84GRR83 pKa = 11.84PEE85 pKa = 3.88RR86 pKa = 11.84GYY88 pKa = 11.11DD89 pKa = 3.48YY90 pKa = 10.71AIKK93 pKa = 10.87DD94 pKa = 3.59GDD96 pKa = 4.02VVAGGLARR104 pKa = 11.84PGGGGLPEE112 pKa = 4.55IANKK116 pKa = 9.29WSEE119 pKa = 4.1IVSAEE124 pKa = 3.84SRR126 pKa = 11.84EE127 pKa = 4.02QFFDD131 pKa = 4.51LLRR134 pKa = 11.84QLDD137 pKa = 4.12PKK139 pKa = 10.46TLVTRR144 pKa = 11.84WTEE147 pKa = 3.7LNKK150 pKa = 10.8YY151 pKa = 10.14ADD153 pKa = 3.7AAYY156 pKa = 10.25APTPEE161 pKa = 5.03PYY163 pKa = 10.25VGSDD167 pKa = 2.91GKK169 pKa = 10.54QFEE172 pKa = 4.79LGMVPEE178 pKa = 4.41LARR181 pKa = 11.84WGEE184 pKa = 3.83QLVANDD190 pKa = 4.12PVEE193 pKa = 4.89GKK195 pKa = 10.38CSPLPGPPRR204 pKa = 11.84GGHH207 pKa = 5.86GGFPGGGPLPLPLALRR223 pKa = 11.84DD224 pKa = 3.5SRR226 pKa = 11.84RR227 pKa = 11.84AMLIVV232 pKa = 3.38
MM1 pKa = 7.47TFLFSARR8 pKa = 11.84YY9 pKa = 9.34VLLTYY14 pKa = 7.24PQCGTLDD21 pKa = 3.23GWAVSDD27 pKa = 4.02HH28 pKa = 6.52LSALGAEE35 pKa = 4.64CIVGRR40 pKa = 11.84EE41 pKa = 4.0NHH43 pKa = 6.3SDD45 pKa = 3.41GGTHH49 pKa = 6.32LHH51 pKa = 6.56AFVDD55 pKa = 4.89FGRR58 pKa = 11.84KK59 pKa = 7.96KK60 pKa = 9.87QSRR63 pKa = 11.84RR64 pKa = 11.84SDD66 pKa = 3.46FFDD69 pKa = 3.73VGGHH73 pKa = 6.35HH74 pKa = 7.14PNIAPSRR81 pKa = 11.84GRR83 pKa = 11.84PEE85 pKa = 3.88RR86 pKa = 11.84GYY88 pKa = 11.11DD89 pKa = 3.48YY90 pKa = 10.71AIKK93 pKa = 10.87DD94 pKa = 3.59GDD96 pKa = 4.02VVAGGLARR104 pKa = 11.84PGGGGLPEE112 pKa = 4.55IANKK116 pKa = 9.29WSEE119 pKa = 4.1IVSAEE124 pKa = 3.84SRR126 pKa = 11.84EE127 pKa = 4.02QFFDD131 pKa = 4.51LLRR134 pKa = 11.84QLDD137 pKa = 4.12PKK139 pKa = 10.46TLVTRR144 pKa = 11.84WTEE147 pKa = 3.7LNKK150 pKa = 10.8YY151 pKa = 10.14ADD153 pKa = 3.7AAYY156 pKa = 10.25APTPEE161 pKa = 5.03PYY163 pKa = 10.25VGSDD167 pKa = 2.91GKK169 pKa = 10.54QFEE172 pKa = 4.79LGMVPEE178 pKa = 4.41LARR181 pKa = 11.84WGEE184 pKa = 3.83QLVANDD190 pKa = 4.12PVEE193 pKa = 4.89GKK195 pKa = 10.38CSPLPGPPRR204 pKa = 11.84GGHH207 pKa = 5.86GGFPGGGPLPLPLALRR223 pKa = 11.84DD224 pKa = 3.5SRR226 pKa = 11.84RR227 pKa = 11.84AMLIVV232 pKa = 3.38
Molecular weight: 25.14 kDa
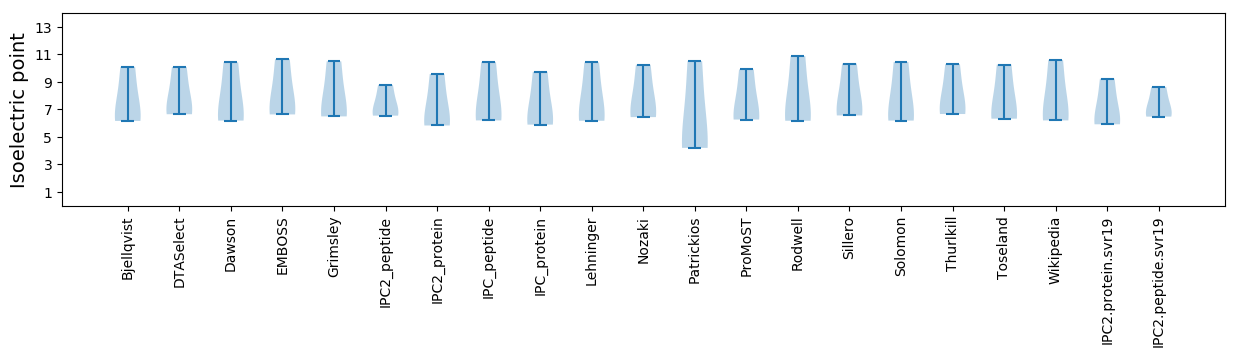
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140CTM1|A0A140CTM1_9CIRC Replication-associated protein OS=Pacific flying fox faeces associated gemycircularvirus-4 OX=1795996 PE=3 SV=1
MM1 pKa = 7.54PLKK4 pKa = 10.21RR5 pKa = 11.84YY6 pKa = 9.5KK7 pKa = 10.66GRR9 pKa = 11.84NSVSYY14 pKa = 10.39SRR16 pKa = 11.84RR17 pKa = 11.84SKK19 pKa = 10.55SRR21 pKa = 11.84KK22 pKa = 8.63PSPKK26 pKa = 8.98KK27 pKa = 10.03KK28 pKa = 10.27SYY30 pKa = 11.04AKK32 pKa = 10.06RR33 pKa = 11.84KK34 pKa = 7.3TMTRR38 pKa = 11.84KK39 pKa = 9.49RR40 pKa = 11.84VVNIASTKK48 pKa = 10.81KK49 pKa = 8.32MDD51 pKa = 3.44TMSTVSIRR59 pKa = 11.84PDD61 pKa = 2.89GSAFRR66 pKa = 11.84EE67 pKa = 4.11PFVMNGDD74 pKa = 3.53TTYY77 pKa = 10.44IIPWVATWRR86 pKa = 11.84NFQTSSTNTVKK97 pKa = 10.64GLKK100 pKa = 9.84ADD102 pKa = 3.88PTRR105 pKa = 11.84SATSCYY111 pKa = 9.37MRR113 pKa = 11.84GLKK116 pKa = 9.62EE117 pKa = 4.03QIQLATNDD125 pKa = 3.86GQCWQWRR132 pKa = 11.84RR133 pKa = 11.84ICFTMKK139 pKa = 10.71GDD141 pKa = 5.11DD142 pKa = 3.39IYY144 pKa = 11.68AAAGPSFRR152 pKa = 11.84VAFQNSGGYY161 pKa = 8.76QRR163 pKa = 11.84LFSDD167 pKa = 3.95WNNTKK172 pKa = 10.63GGTTPPINFLVDD184 pKa = 4.3PIFKK188 pKa = 8.96GTAGTDD194 pKa = 2.77WDD196 pKa = 4.68DD197 pKa = 4.15PFVAPLDD204 pKa = 3.89PYY206 pKa = 10.85RR207 pKa = 11.84IGVKK211 pKa = 9.67YY212 pKa = 10.89DD213 pKa = 2.84KK214 pKa = 10.43TRR216 pKa = 11.84YY217 pKa = 8.9IRR219 pKa = 11.84SGNNFGMLKK228 pKa = 9.2MVKK231 pKa = 9.52LWHH234 pKa = 6.71PMNKK238 pKa = 8.34TLIYY242 pKa = 10.77NDD244 pKa = 4.48DD245 pKa = 3.89EE246 pKa = 5.3NGGVTDD252 pKa = 4.56AAATSVRR259 pKa = 11.84THH261 pKa = 5.79GMGDD265 pKa = 3.71YY266 pKa = 10.82YY267 pKa = 11.11VIDD270 pKa = 3.93MFRR273 pKa = 11.84AATGAVAGSQLNIRR287 pKa = 11.84SSGG290 pKa = 3.33
MM1 pKa = 7.54PLKK4 pKa = 10.21RR5 pKa = 11.84YY6 pKa = 9.5KK7 pKa = 10.66GRR9 pKa = 11.84NSVSYY14 pKa = 10.39SRR16 pKa = 11.84RR17 pKa = 11.84SKK19 pKa = 10.55SRR21 pKa = 11.84KK22 pKa = 8.63PSPKK26 pKa = 8.98KK27 pKa = 10.03KK28 pKa = 10.27SYY30 pKa = 11.04AKK32 pKa = 10.06RR33 pKa = 11.84KK34 pKa = 7.3TMTRR38 pKa = 11.84KK39 pKa = 9.49RR40 pKa = 11.84VVNIASTKK48 pKa = 10.81KK49 pKa = 8.32MDD51 pKa = 3.44TMSTVSIRR59 pKa = 11.84PDD61 pKa = 2.89GSAFRR66 pKa = 11.84EE67 pKa = 4.11PFVMNGDD74 pKa = 3.53TTYY77 pKa = 10.44IIPWVATWRR86 pKa = 11.84NFQTSSTNTVKK97 pKa = 10.64GLKK100 pKa = 9.84ADD102 pKa = 3.88PTRR105 pKa = 11.84SATSCYY111 pKa = 9.37MRR113 pKa = 11.84GLKK116 pKa = 9.62EE117 pKa = 4.03QIQLATNDD125 pKa = 3.86GQCWQWRR132 pKa = 11.84RR133 pKa = 11.84ICFTMKK139 pKa = 10.71GDD141 pKa = 5.11DD142 pKa = 3.39IYY144 pKa = 11.68AAAGPSFRR152 pKa = 11.84VAFQNSGGYY161 pKa = 8.76QRR163 pKa = 11.84LFSDD167 pKa = 3.95WNNTKK172 pKa = 10.63GGTTPPINFLVDD184 pKa = 4.3PIFKK188 pKa = 8.96GTAGTDD194 pKa = 2.77WDD196 pKa = 4.68DD197 pKa = 4.15PFVAPLDD204 pKa = 3.89PYY206 pKa = 10.85RR207 pKa = 11.84IGVKK211 pKa = 9.67YY212 pKa = 10.89DD213 pKa = 2.84KK214 pKa = 10.43TRR216 pKa = 11.84YY217 pKa = 8.9IRR219 pKa = 11.84SGNNFGMLKK228 pKa = 9.2MVKK231 pKa = 9.52LWHH234 pKa = 6.71PMNKK238 pKa = 8.34TLIYY242 pKa = 10.77NDD244 pKa = 4.48DD245 pKa = 3.89EE246 pKa = 5.3NGGVTDD252 pKa = 4.56AAATSVRR259 pKa = 11.84THH261 pKa = 5.79GMGDD265 pKa = 3.71YY266 pKa = 10.82YY267 pKa = 11.11VIDD270 pKa = 3.93MFRR273 pKa = 11.84AATGAVAGSQLNIRR287 pKa = 11.84SSGG290 pKa = 3.33
Molecular weight: 32.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
858 |
232 |
336 |
286.0 |
31.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.809 ± 0.348 | 1.166 ± 0.05 |
6.41 ± 0.078 | 3.963 ± 1.084 |
4.429 ± 0.162 | 10.606 ± 0.956 |
1.981 ± 0.476 | 3.263 ± 0.595 |
5.128 ± 1.033 | 7.809 ± 1.496 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.214 ± 0.723 | 3.613 ± 0.7 |
6.41 ± 0.684 | 2.681 ± 0.032 |
7.459 ± 0.189 | 6.41 ± 0.566 |
5.828 ± 1.152 | 6.177 ± 0.333 |
2.681 ± 0.446 | 3.963 ± 0.292 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
