
Cottoperca gobio (Frogmouth) (Aphritis gobio)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii; Actinopteri; Neopterygii; Teleostei; Osteoglossocephalai; Clupeocephala; Euteleosteomorpha; Neoteleostei; Eurypterygia; Ctenosquamata;
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins
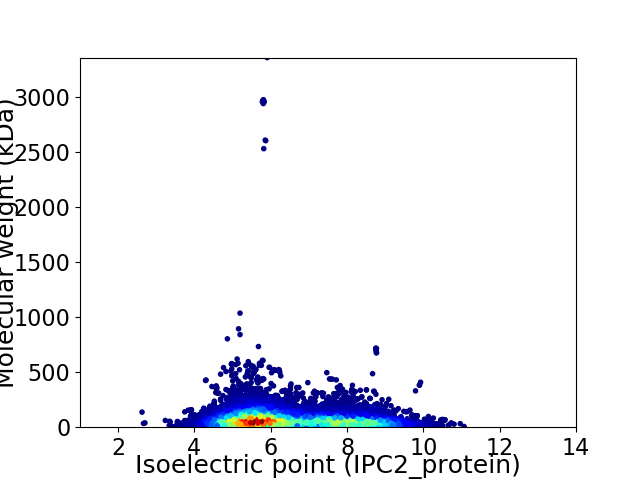
Virtual 2D-PAGE plot for 32862 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6J2PBQ2|A0A6J2PBQ2_COTGO src substrate cortactin isoform X3 OS=Cottoperca gobio OX=56716 GN=cttn PE=4 SV=1
SS1 pKa = 6.52QDD3 pKa = 3.59CVDD6 pKa = 3.83IDD8 pKa = 3.94EE9 pKa = 5.6CVDD12 pKa = 4.14LPDD15 pKa = 4.03ACVSHH20 pKa = 6.05SVCINTVGSYY30 pKa = 10.42KK31 pKa = 10.7CGGCKK36 pKa = 9.77PGFLGNQTSGCLPRR50 pKa = 11.84KK51 pKa = 9.62SCAALTFNPCDD62 pKa = 3.71INAHH66 pKa = 5.22CTMEE70 pKa = 4.09RR71 pKa = 11.84SGEE74 pKa = 4.19VACRR78 pKa = 11.84CNVGWAGNGNTCGTDD93 pKa = 3.18TDD95 pKa = 3.49IDD97 pKa = 4.4GYY99 pKa = 9.25PNRR102 pKa = 11.84SLPCMDD108 pKa = 4.32NDD110 pKa = 3.55KK111 pKa = 10.97HH112 pKa = 6.0GQTGQLCVHH121 pKa = 6.63PHH123 pKa = 6.18SGQEE127 pKa = 3.99DD128 pKa = 4.05ADD130 pKa = 3.85GDD132 pKa = 4.58GIGDD136 pKa = 3.76QCDD139 pKa = 3.33EE140 pKa = 4.81DD141 pKa = 5.57ADD143 pKa = 4.08GDD145 pKa = 4.34GIKK148 pKa = 10.51NVEE151 pKa = 4.35DD152 pKa = 3.49NCRR155 pKa = 11.84LVPNKK160 pKa = 10.07DD161 pKa = 3.51QQNSDD166 pKa = 3.18SDD168 pKa = 4.33SFGDD172 pKa = 4.74ACDD175 pKa = 3.62NCPNVPNADD184 pKa = 3.72QKK186 pKa = 11.15DD187 pKa = 3.81TDD189 pKa = 4.29SNGQGDD195 pKa = 4.7ACDD198 pKa = 3.56QDD200 pKa = 3.22IDD202 pKa = 4.35GDD204 pKa = 4.4GIPNVLDD211 pKa = 3.65NCPKK215 pKa = 10.25VPNPMQTDD223 pKa = 3.39RR224 pKa = 11.84DD225 pKa = 3.68RR226 pKa = 11.84DD227 pKa = 4.03GVGDD231 pKa = 4.08ACDD234 pKa = 3.57SCPEE238 pKa = 4.52LSNPMQTDD246 pKa = 2.72IDD248 pKa = 3.96NDD250 pKa = 4.08LVGDD254 pKa = 3.98VCDD257 pKa = 4.47TNQDD261 pKa = 3.05MDD263 pKa = 4.19GDD265 pKa = 4.35GLQDD269 pKa = 4.49SRR271 pKa = 11.84DD272 pKa = 3.79NCPDD276 pKa = 3.19IPNSSQLDD284 pKa = 3.58SDD286 pKa = 4.01NDD288 pKa = 4.11GLGDD292 pKa = 5.08DD293 pKa = 5.35CDD295 pKa = 5.44HH296 pKa = 7.79DD297 pKa = 4.92DD298 pKa = 5.57DD299 pKa = 5.8NDD301 pKa = 4.14GVLDD305 pKa = 5.21DD306 pKa = 4.8YY307 pKa = 11.88DD308 pKa = 3.61NCRR311 pKa = 11.84LVVNPNQKK319 pKa = 10.58DD320 pKa = 3.17SDD322 pKa = 4.05MNGVGDD328 pKa = 3.64VCEE331 pKa = 4.3NDD333 pKa = 3.42FDD335 pKa = 4.38NDD337 pKa = 3.56AVMDD341 pKa = 4.47LVDD344 pKa = 4.57ACPEE348 pKa = 4.08SAEE351 pKa = 4.1VTLTDD356 pKa = 3.82FRR358 pKa = 11.84AYY360 pKa = 8.36QTVILDD366 pKa = 3.91PEE368 pKa = 5.2GDD370 pKa = 3.72AQIDD374 pKa = 4.0PTWVVLNQGMEE385 pKa = 4.11IVQTMNSDD393 pKa = 3.19PGLAVGYY400 pKa = 7.31TAFNGVDD407 pKa = 3.99FEE409 pKa = 4.97GTFHH413 pKa = 6.81VNTVTDD419 pKa = 3.57DD420 pKa = 4.08DD421 pKa = 4.35YY422 pKa = 12.0AGFIFGYY429 pKa = 9.58QDD431 pKa = 2.83SSSFYY436 pKa = 10.47VVMWKK441 pKa = 7.78QTEE444 pKa = 3.81QTYY447 pKa = 8.19WQSLPFRR454 pKa = 11.84AAAQPALQLKK464 pKa = 9.2AVKK467 pKa = 10.3SRR469 pKa = 11.84TGPGEE474 pKa = 3.87FLRR477 pKa = 11.84NALWHH482 pKa = 6.33TGDD485 pKa = 3.39TPGEE489 pKa = 4.12VKK491 pKa = 10.69LLWKK495 pKa = 10.41DD496 pKa = 3.35PRR498 pKa = 11.84NVGWKK503 pKa = 10.34DD504 pKa = 3.01KK505 pKa = 10.86TSYY508 pKa = 10.19RR509 pKa = 11.84WQLSHH514 pKa = 7.03RR515 pKa = 11.84PQVGYY520 pKa = 9.37MRR522 pKa = 11.84VKK524 pKa = 10.64LFEE527 pKa = 4.36GTDD530 pKa = 3.6LVADD534 pKa = 3.84SGVVIDD540 pKa = 3.46TTMRR544 pKa = 11.84GGRR547 pKa = 11.84LGVFCFSQEE556 pKa = 3.94NIIWSNLRR564 pKa = 11.84YY565 pKa = 9.76RR566 pKa = 11.84CNDD569 pKa = 3.41TLPEE573 pKa = 4.29DD574 pKa = 4.26FQTHH578 pKa = 5.09RR579 pKa = 11.84QQFLMHH585 pKa = 6.27IRR587 pKa = 11.84VV588 pKa = 3.36
SS1 pKa = 6.52QDD3 pKa = 3.59CVDD6 pKa = 3.83IDD8 pKa = 3.94EE9 pKa = 5.6CVDD12 pKa = 4.14LPDD15 pKa = 4.03ACVSHH20 pKa = 6.05SVCINTVGSYY30 pKa = 10.42KK31 pKa = 10.7CGGCKK36 pKa = 9.77PGFLGNQTSGCLPRR50 pKa = 11.84KK51 pKa = 9.62SCAALTFNPCDD62 pKa = 3.71INAHH66 pKa = 5.22CTMEE70 pKa = 4.09RR71 pKa = 11.84SGEE74 pKa = 4.19VACRR78 pKa = 11.84CNVGWAGNGNTCGTDD93 pKa = 3.18TDD95 pKa = 3.49IDD97 pKa = 4.4GYY99 pKa = 9.25PNRR102 pKa = 11.84SLPCMDD108 pKa = 4.32NDD110 pKa = 3.55KK111 pKa = 10.97HH112 pKa = 6.0GQTGQLCVHH121 pKa = 6.63PHH123 pKa = 6.18SGQEE127 pKa = 3.99DD128 pKa = 4.05ADD130 pKa = 3.85GDD132 pKa = 4.58GIGDD136 pKa = 3.76QCDD139 pKa = 3.33EE140 pKa = 4.81DD141 pKa = 5.57ADD143 pKa = 4.08GDD145 pKa = 4.34GIKK148 pKa = 10.51NVEE151 pKa = 4.35DD152 pKa = 3.49NCRR155 pKa = 11.84LVPNKK160 pKa = 10.07DD161 pKa = 3.51QQNSDD166 pKa = 3.18SDD168 pKa = 4.33SFGDD172 pKa = 4.74ACDD175 pKa = 3.62NCPNVPNADD184 pKa = 3.72QKK186 pKa = 11.15DD187 pKa = 3.81TDD189 pKa = 4.29SNGQGDD195 pKa = 4.7ACDD198 pKa = 3.56QDD200 pKa = 3.22IDD202 pKa = 4.35GDD204 pKa = 4.4GIPNVLDD211 pKa = 3.65NCPKK215 pKa = 10.25VPNPMQTDD223 pKa = 3.39RR224 pKa = 11.84DD225 pKa = 3.68RR226 pKa = 11.84DD227 pKa = 4.03GVGDD231 pKa = 4.08ACDD234 pKa = 3.57SCPEE238 pKa = 4.52LSNPMQTDD246 pKa = 2.72IDD248 pKa = 3.96NDD250 pKa = 4.08LVGDD254 pKa = 3.98VCDD257 pKa = 4.47TNQDD261 pKa = 3.05MDD263 pKa = 4.19GDD265 pKa = 4.35GLQDD269 pKa = 4.49SRR271 pKa = 11.84DD272 pKa = 3.79NCPDD276 pKa = 3.19IPNSSQLDD284 pKa = 3.58SDD286 pKa = 4.01NDD288 pKa = 4.11GLGDD292 pKa = 5.08DD293 pKa = 5.35CDD295 pKa = 5.44HH296 pKa = 7.79DD297 pKa = 4.92DD298 pKa = 5.57DD299 pKa = 5.8NDD301 pKa = 4.14GVLDD305 pKa = 5.21DD306 pKa = 4.8YY307 pKa = 11.88DD308 pKa = 3.61NCRR311 pKa = 11.84LVVNPNQKK319 pKa = 10.58DD320 pKa = 3.17SDD322 pKa = 4.05MNGVGDD328 pKa = 3.64VCEE331 pKa = 4.3NDD333 pKa = 3.42FDD335 pKa = 4.38NDD337 pKa = 3.56AVMDD341 pKa = 4.47LVDD344 pKa = 4.57ACPEE348 pKa = 4.08SAEE351 pKa = 4.1VTLTDD356 pKa = 3.82FRR358 pKa = 11.84AYY360 pKa = 8.36QTVILDD366 pKa = 3.91PEE368 pKa = 5.2GDD370 pKa = 3.72AQIDD374 pKa = 4.0PTWVVLNQGMEE385 pKa = 4.11IVQTMNSDD393 pKa = 3.19PGLAVGYY400 pKa = 7.31TAFNGVDD407 pKa = 3.99FEE409 pKa = 4.97GTFHH413 pKa = 6.81VNTVTDD419 pKa = 3.57DD420 pKa = 4.08DD421 pKa = 4.35YY422 pKa = 12.0AGFIFGYY429 pKa = 9.58QDD431 pKa = 2.83SSSFYY436 pKa = 10.47VVMWKK441 pKa = 7.78QTEE444 pKa = 3.81QTYY447 pKa = 8.19WQSLPFRR454 pKa = 11.84AAAQPALQLKK464 pKa = 9.2AVKK467 pKa = 10.3SRR469 pKa = 11.84TGPGEE474 pKa = 3.87FLRR477 pKa = 11.84NALWHH482 pKa = 6.33TGDD485 pKa = 3.39TPGEE489 pKa = 4.12VKK491 pKa = 10.69LLWKK495 pKa = 10.41DD496 pKa = 3.35PRR498 pKa = 11.84NVGWKK503 pKa = 10.34DD504 pKa = 3.01KK505 pKa = 10.86TSYY508 pKa = 10.19RR509 pKa = 11.84WQLSHH514 pKa = 7.03RR515 pKa = 11.84PQVGYY520 pKa = 9.37MRR522 pKa = 11.84VKK524 pKa = 10.64LFEE527 pKa = 4.36GTDD530 pKa = 3.6LVADD534 pKa = 3.84SGVVIDD540 pKa = 3.46TTMRR544 pKa = 11.84GGRR547 pKa = 11.84LGVFCFSQEE556 pKa = 3.94NIIWSNLRR564 pKa = 11.84YY565 pKa = 9.76RR566 pKa = 11.84CNDD569 pKa = 3.41TLPEE573 pKa = 4.29DD574 pKa = 4.26FQTHH578 pKa = 5.09RR579 pKa = 11.84QQFLMHH585 pKa = 6.27IRR587 pKa = 11.84VV588 pKa = 3.36
Molecular weight: 64.38 kDa
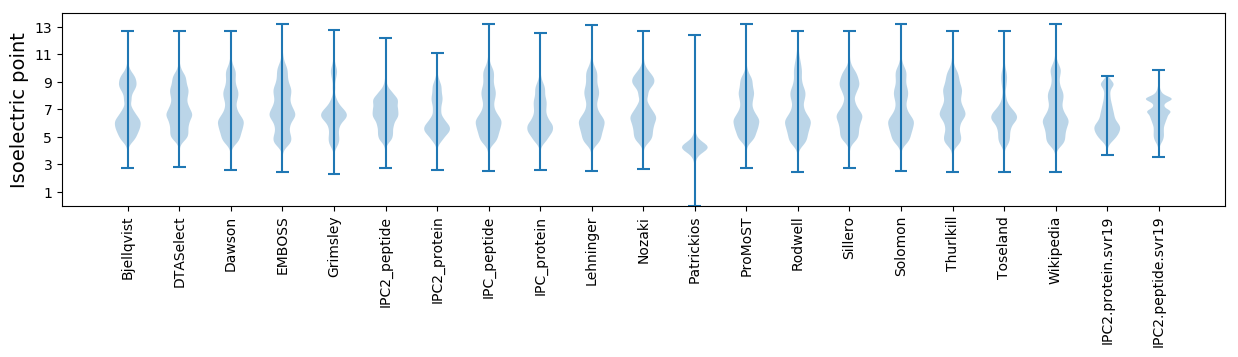
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6J2QMP6|A0A6J2QMP6_COTGO protein FAM131B-like isoform X1 OS=Cottoperca gobio OX=56716 GN=LOC115015785 PE=3 SV=1
MM1 pKa = 7.12MMKK4 pKa = 9.6ICVLLLLCVASGNTQITVAPTTAAPAAATTVAPTTAAPAAATTVAPTTAAPAAATTAAPPATTAAPAAITAEE76 pKa = 4.18ATTAAPAAITAEE88 pKa = 4.18ATTAAPAAITAEE100 pKa = 4.19ATTVAPAAITAATAAATTAAAPAAITAATAAATTAAAPAATTAAATVAPAATATAAAATTVAPTTAAPAQQPQLPQPQRR179 pKa = 11.84PRR181 pKa = 11.84QQQPRR186 pKa = 11.84RR187 pKa = 11.84PRR189 pKa = 11.84LQLQRR194 pKa = 11.84TQPRR198 pKa = 11.84RR199 pKa = 11.84PRR201 pKa = 11.84QQLQRR206 pKa = 11.84KK207 pKa = 5.87QPRR210 pKa = 11.84RR211 pKa = 11.84PRR213 pKa = 11.84QQLQRR218 pKa = 11.84KK219 pKa = 5.87QPRR222 pKa = 11.84RR223 pKa = 11.84PRR225 pKa = 11.84QQLQRR230 pKa = 11.84KK231 pKa = 5.87QPRR234 pKa = 11.84RR235 pKa = 11.84PRR237 pKa = 11.84QQLQRR242 pKa = 11.84KK243 pKa = 5.01QPRR246 pKa = 11.84RR247 pKa = 11.84RR248 pKa = 11.84RR249 pKa = 11.84QQLQRR254 pKa = 11.84KK255 pKa = 5.88QPRR258 pKa = 11.84RR259 pKa = 11.84PRR261 pKa = 11.84QQLQRR266 pKa = 11.84KK267 pKa = 7.41QPQRR271 pKa = 11.84PRR273 pKa = 11.84QQPRR277 pKa = 11.84RR278 pKa = 11.84KK279 pKa = 8.59PRR281 pKa = 11.84RR282 pKa = 11.84PRR284 pKa = 11.84HH285 pKa = 5.06HH286 pKa = 6.55QQLQLPPLQSSTKK299 pKa = 9.62QVSFF303 pKa = 4.06
MM1 pKa = 7.12MMKK4 pKa = 9.6ICVLLLLCVASGNTQITVAPTTAAPAAATTVAPTTAAPAAATTVAPTTAAPAAATTAAPPATTAAPAAITAEE76 pKa = 4.18ATTAAPAAITAEE88 pKa = 4.18ATTAAPAAITAEE100 pKa = 4.19ATTVAPAAITAATAAATTAAAPAAITAATAAATTAAAPAATTAAATVAPAATATAAAATTVAPTTAAPAQQPQLPQPQRR179 pKa = 11.84PRR181 pKa = 11.84QQQPRR186 pKa = 11.84RR187 pKa = 11.84PRR189 pKa = 11.84LQLQRR194 pKa = 11.84TQPRR198 pKa = 11.84RR199 pKa = 11.84PRR201 pKa = 11.84QQLQRR206 pKa = 11.84KK207 pKa = 5.87QPRR210 pKa = 11.84RR211 pKa = 11.84PRR213 pKa = 11.84QQLQRR218 pKa = 11.84KK219 pKa = 5.87QPRR222 pKa = 11.84RR223 pKa = 11.84PRR225 pKa = 11.84QQLQRR230 pKa = 11.84KK231 pKa = 5.87QPRR234 pKa = 11.84RR235 pKa = 11.84PRR237 pKa = 11.84QQLQRR242 pKa = 11.84KK243 pKa = 5.01QPRR246 pKa = 11.84RR247 pKa = 11.84RR248 pKa = 11.84RR249 pKa = 11.84QQLQRR254 pKa = 11.84KK255 pKa = 5.88QPRR258 pKa = 11.84RR259 pKa = 11.84PRR261 pKa = 11.84QQLQRR266 pKa = 11.84KK267 pKa = 7.41QPQRR271 pKa = 11.84PRR273 pKa = 11.84QQPRR277 pKa = 11.84RR278 pKa = 11.84KK279 pKa = 8.59PRR281 pKa = 11.84RR282 pKa = 11.84PRR284 pKa = 11.84HH285 pKa = 5.06HH286 pKa = 6.55QQLQLPPLQSSTKK299 pKa = 9.62QVSFF303 pKa = 4.06
Molecular weight: 32.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
22561719 |
31 |
30276 |
686.6 |
76.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.61 ± 0.011 | 2.109 ± 0.01 |
5.273 ± 0.008 | 7.038 ± 0.017 |
3.339 ± 0.01 | 6.373 ± 0.02 |
2.663 ± 0.009 | 4.236 ± 0.013 |
5.673 ± 0.019 | 9.237 ± 0.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.397 ± 0.006 | 3.797 ± 0.009 |
5.943 ± 0.02 | 4.904 ± 0.022 |
5.68 ± 0.011 | 8.98 ± 0.02 |
5.759 ± 0.015 | 6.26 ± 0.02 |
1.087 ± 0.005 | 2.64 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
