
Streptococcus satellite phage Javan581
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.74
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 17 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4D5ZRR8|A0A4D5ZRR8_9VIRU HTH cro/C1-type domain-containing protein OS=Streptococcus satellite phage Javan581 OX=2558757 GN=JavanS581_0014 PE=4 SV=1
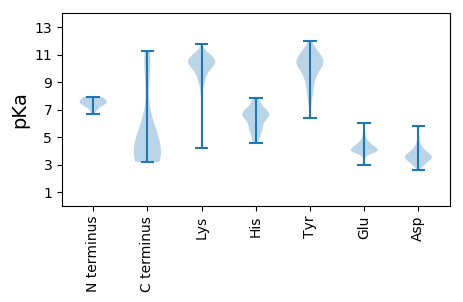
MM1 pKa = 7.67TDD3 pKa = 3.07DD4 pKa = 3.59TEE6 pKa = 4.18IRR8 pKa = 11.84KK9 pKa = 8.93LFQIYY14 pKa = 7.31QTTRR18 pKa = 11.84EE19 pKa = 4.26NKK21 pKa = 10.38DD22 pKa = 2.94MTLNTFSDD30 pKa = 3.76TANTFTFNYY39 pKa = 8.15TFKK42 pKa = 11.11DD43 pKa = 3.04HH44 pKa = 7.52DD45 pKa = 3.88TAQVAGHH52 pKa = 6.91ALMGYY57 pKa = 6.39MTGTFEE63 pKa = 4.03QPGIEE68 pKa = 4.09VYY70 pKa = 10.97YY71 pKa = 11.08DD72 pKa = 3.31NDD74 pKa = 4.04KK75 pKa = 11.67VGGDD79 pKa = 3.8YY80 pKa = 11.12NRR82 pKa = 11.84LAVEE86 pKa = 4.09YY87 pKa = 10.78VADD90 pKa = 4.0TEE92 pKa = 4.19LTEE95 pKa = 4.12TFRR98 pKa = 11.84RR99 pKa = 11.84ICDD102 pKa = 3.58SFQGYY107 pKa = 10.2YY108 pKa = 10.3NDD110 pKa = 4.91PEE112 pKa = 6.04AEE114 pKa = 4.06TDD116 pKa = 3.75VEE118 pKa = 4.02DD119 pKa = 3.49QYY121 pKa = 11.87RR122 pKa = 11.84LEE124 pKa = 4.22RR125 pKa = 11.84VEE127 pKa = 3.91QLKK130 pKa = 10.42QSEE133 pKa = 4.72TFDD136 pKa = 3.59SLLEE140 pKa = 3.94KK141 pKa = 10.66VVIYY145 pKa = 10.18EE146 pKa = 5.3LEE148 pKa = 4.06LLDD151 pKa = 3.69YY152 pKa = 11.22AEE154 pKa = 5.48RR155 pKa = 11.84LLSDD159 pKa = 4.65DD160 pKa = 5.61PIPTDD165 pKa = 3.09TEE167 pKa = 3.97MAYY170 pKa = 7.69MTLNLIGGKK179 pKa = 10.05GVGLFKK185 pKa = 11.15SLDD188 pKa = 3.51EE189 pKa = 4.84DD190 pKa = 3.81NEE192 pKa = 4.35YY193 pKa = 11.31SGLAYY198 pKa = 10.92YY199 pKa = 10.23NAEE202 pKa = 4.13AEE204 pKa = 4.28
MM1 pKa = 7.67TDD3 pKa = 3.07DD4 pKa = 3.59TEE6 pKa = 4.18IRR8 pKa = 11.84KK9 pKa = 8.93LFQIYY14 pKa = 7.31QTTRR18 pKa = 11.84EE19 pKa = 4.26NKK21 pKa = 10.38DD22 pKa = 2.94MTLNTFSDD30 pKa = 3.76TANTFTFNYY39 pKa = 8.15TFKK42 pKa = 11.11DD43 pKa = 3.04HH44 pKa = 7.52DD45 pKa = 3.88TAQVAGHH52 pKa = 6.91ALMGYY57 pKa = 6.39MTGTFEE63 pKa = 4.03QPGIEE68 pKa = 4.09VYY70 pKa = 10.97YY71 pKa = 11.08DD72 pKa = 3.31NDD74 pKa = 4.04KK75 pKa = 11.67VGGDD79 pKa = 3.8YY80 pKa = 11.12NRR82 pKa = 11.84LAVEE86 pKa = 4.09YY87 pKa = 10.78VADD90 pKa = 4.0TEE92 pKa = 4.19LTEE95 pKa = 4.12TFRR98 pKa = 11.84RR99 pKa = 11.84ICDD102 pKa = 3.58SFQGYY107 pKa = 10.2YY108 pKa = 10.3NDD110 pKa = 4.91PEE112 pKa = 6.04AEE114 pKa = 4.06TDD116 pKa = 3.75VEE118 pKa = 4.02DD119 pKa = 3.49QYY121 pKa = 11.87RR122 pKa = 11.84LEE124 pKa = 4.22RR125 pKa = 11.84VEE127 pKa = 3.91QLKK130 pKa = 10.42QSEE133 pKa = 4.72TFDD136 pKa = 3.59SLLEE140 pKa = 3.94KK141 pKa = 10.66VVIYY145 pKa = 10.18EE146 pKa = 5.3LEE148 pKa = 4.06LLDD151 pKa = 3.69YY152 pKa = 11.22AEE154 pKa = 5.48RR155 pKa = 11.84LLSDD159 pKa = 4.65DD160 pKa = 5.61PIPTDD165 pKa = 3.09TEE167 pKa = 3.97MAYY170 pKa = 7.69MTLNLIGGKK179 pKa = 10.05GVGLFKK185 pKa = 11.15SLDD188 pKa = 3.51EE189 pKa = 4.84DD190 pKa = 3.81NEE192 pKa = 4.35YY193 pKa = 11.31SGLAYY198 pKa = 10.92YY199 pKa = 10.23NAEE202 pKa = 4.13AEE204 pKa = 4.28
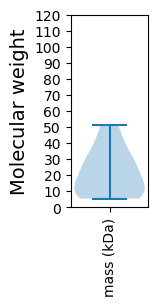
Molecular weight: 23.56 kDa
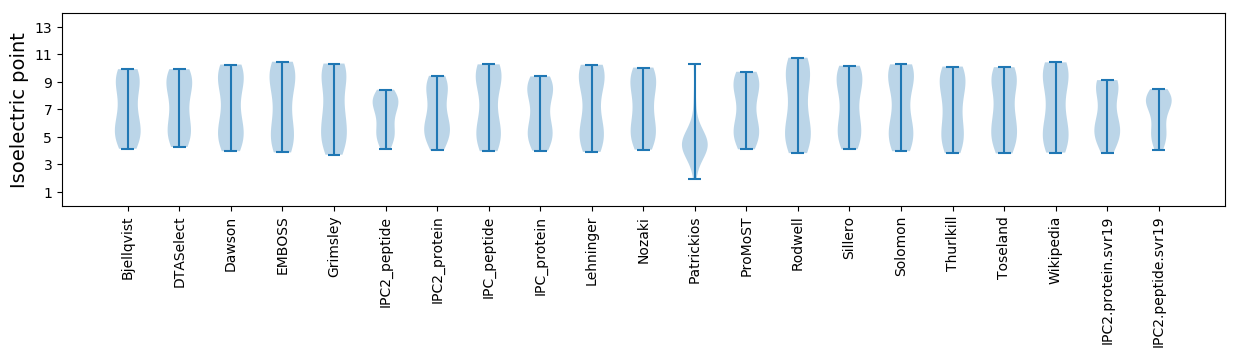
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D5ZR50|A0A4D5ZR50_9VIRU Uncharacterized protein OS=Streptococcus satellite phage Javan581 OX=2558757 GN=JavanS581_0005 PE=4 SV=1
MM1 pKa = 7.38IMKK4 pKa = 8.9ITEE7 pKa = 4.06VKK9 pKa = 9.83KK10 pKa = 10.92KK11 pKa = 10.69NGATVYY17 pKa = 10.17RR18 pKa = 11.84ASVYY22 pKa = 10.76LGIDD26 pKa = 4.16AITGKK31 pKa = 10.19KK32 pKa = 10.01VKK34 pKa = 10.03TNVTGRR40 pKa = 11.84TKK42 pKa = 10.82KK43 pKa = 9.46EE44 pKa = 4.12VKK46 pKa = 10.43NKK48 pKa = 6.58TQQAIATFKK57 pKa = 10.34TDD59 pKa = 2.86GATRR63 pKa = 11.84YY64 pKa = 9.71QNATITSYY72 pKa = 11.5KK73 pKa = 10.12EE74 pKa = 4.02LAEE77 pKa = 4.25LWWNSYY83 pKa = 8.4KK84 pKa = 9.73HH85 pKa = 4.85TVKK88 pKa = 10.64PNTRR92 pKa = 11.84GNIRR96 pKa = 11.84GLLKK100 pKa = 10.74NHH102 pKa = 6.69VIPLFGAYY110 pKa = 9.51KK111 pKa = 10.15LDD113 pKa = 3.71KK114 pKa = 10.24LTTPLLQSIVIKK126 pKa = 10.71LADD129 pKa = 3.5KK130 pKa = 11.26ANTGEE135 pKa = 4.04AGAYY139 pKa = 8.96LHH141 pKa = 6.56YY142 pKa = 11.05DD143 pKa = 4.09KK144 pKa = 11.05IHH146 pKa = 6.58ALNKK150 pKa = 10.18RR151 pKa = 11.84ILQYY155 pKa = 10.87GVVMQVLPYY164 pKa = 9.81NPARR168 pKa = 11.84EE169 pKa = 4.25VILPRR174 pKa = 11.84NAKK177 pKa = 9.33KK178 pKa = 9.43ATRR181 pKa = 11.84KK182 pKa = 9.25KK183 pKa = 10.03VKK185 pKa = 10.16HH186 pKa = 6.24FNDD189 pKa = 3.34EE190 pKa = 3.86QLKK193 pKa = 10.06QFLGYY198 pKa = 10.59LDD200 pKa = 5.41GLDD203 pKa = 3.24LTNYY207 pKa = 9.51RR208 pKa = 11.84NLYY211 pKa = 9.16EE212 pKa = 3.97ATLYY216 pKa = 10.98KK217 pKa = 10.43FLLATGCRR225 pKa = 11.84INEE228 pKa = 3.89VLALHH233 pKa = 6.87WSDD236 pKa = 4.12IDD238 pKa = 4.14LNNATVSITKK248 pKa = 8.91TLNRR252 pKa = 11.84YY253 pKa = 9.22GSINSPKK260 pKa = 9.97SNASIRR266 pKa = 11.84DD267 pKa = 3.53INIDD271 pKa = 3.67SQTVAMMKK279 pKa = 9.85EE280 pKa = 3.79YY281 pKa = 10.34RR282 pKa = 11.84RR283 pKa = 11.84RR284 pKa = 11.84QIQEE288 pKa = 2.98AWTLGRR294 pKa = 11.84SEE296 pKa = 4.42TVVFSDD302 pKa = 5.82FIHH305 pKa = 7.8DD306 pKa = 3.96YY307 pKa = 11.31PEE309 pKa = 5.01DD310 pKa = 3.47KK311 pKa = 10.71TLGNRR316 pKa = 11.84LTTRR320 pKa = 11.84LRR322 pKa = 11.84NIGLPNIGFHH332 pKa = 6.38GFRR335 pKa = 11.84HH336 pKa = 4.87THH338 pKa = 6.58ASLLLNSGIPYY349 pKa = 10.0KK350 pKa = 10.34EE351 pKa = 3.8LQHH354 pKa = 6.73RR355 pKa = 11.84LGHH358 pKa = 5.38SRR360 pKa = 11.84ISITMDD366 pKa = 2.92IYY368 pKa = 11.54SHH370 pKa = 7.04LSKK373 pKa = 10.96EE374 pKa = 4.11NAKK377 pKa = 10.48NAVAFYY383 pKa = 10.59EE384 pKa = 4.29KK385 pKa = 10.97ALGNLL390 pKa = 3.69
MM1 pKa = 7.38IMKK4 pKa = 8.9ITEE7 pKa = 4.06VKK9 pKa = 9.83KK10 pKa = 10.92KK11 pKa = 10.69NGATVYY17 pKa = 10.17RR18 pKa = 11.84ASVYY22 pKa = 10.76LGIDD26 pKa = 4.16AITGKK31 pKa = 10.19KK32 pKa = 10.01VKK34 pKa = 10.03TNVTGRR40 pKa = 11.84TKK42 pKa = 10.82KK43 pKa = 9.46EE44 pKa = 4.12VKK46 pKa = 10.43NKK48 pKa = 6.58TQQAIATFKK57 pKa = 10.34TDD59 pKa = 2.86GATRR63 pKa = 11.84YY64 pKa = 9.71QNATITSYY72 pKa = 11.5KK73 pKa = 10.12EE74 pKa = 4.02LAEE77 pKa = 4.25LWWNSYY83 pKa = 8.4KK84 pKa = 9.73HH85 pKa = 4.85TVKK88 pKa = 10.64PNTRR92 pKa = 11.84GNIRR96 pKa = 11.84GLLKK100 pKa = 10.74NHH102 pKa = 6.69VIPLFGAYY110 pKa = 9.51KK111 pKa = 10.15LDD113 pKa = 3.71KK114 pKa = 10.24LTTPLLQSIVIKK126 pKa = 10.71LADD129 pKa = 3.5KK130 pKa = 11.26ANTGEE135 pKa = 4.04AGAYY139 pKa = 8.96LHH141 pKa = 6.56YY142 pKa = 11.05DD143 pKa = 4.09KK144 pKa = 11.05IHH146 pKa = 6.58ALNKK150 pKa = 10.18RR151 pKa = 11.84ILQYY155 pKa = 10.87GVVMQVLPYY164 pKa = 9.81NPARR168 pKa = 11.84EE169 pKa = 4.25VILPRR174 pKa = 11.84NAKK177 pKa = 9.33KK178 pKa = 9.43ATRR181 pKa = 11.84KK182 pKa = 9.25KK183 pKa = 10.03VKK185 pKa = 10.16HH186 pKa = 6.24FNDD189 pKa = 3.34EE190 pKa = 3.86QLKK193 pKa = 10.06QFLGYY198 pKa = 10.59LDD200 pKa = 5.41GLDD203 pKa = 3.24LTNYY207 pKa = 9.51RR208 pKa = 11.84NLYY211 pKa = 9.16EE212 pKa = 3.97ATLYY216 pKa = 10.98KK217 pKa = 10.43FLLATGCRR225 pKa = 11.84INEE228 pKa = 3.89VLALHH233 pKa = 6.87WSDD236 pKa = 4.12IDD238 pKa = 4.14LNNATVSITKK248 pKa = 8.91TLNRR252 pKa = 11.84YY253 pKa = 9.22GSINSPKK260 pKa = 9.97SNASIRR266 pKa = 11.84DD267 pKa = 3.53INIDD271 pKa = 3.67SQTVAMMKK279 pKa = 9.85EE280 pKa = 3.79YY281 pKa = 10.34RR282 pKa = 11.84RR283 pKa = 11.84RR284 pKa = 11.84QIQEE288 pKa = 2.98AWTLGRR294 pKa = 11.84SEE296 pKa = 4.42TVVFSDD302 pKa = 5.82FIHH305 pKa = 7.8DD306 pKa = 3.96YY307 pKa = 11.31PEE309 pKa = 5.01DD310 pKa = 3.47KK311 pKa = 10.71TLGNRR316 pKa = 11.84LTTRR320 pKa = 11.84LRR322 pKa = 11.84NIGLPNIGFHH332 pKa = 6.38GFRR335 pKa = 11.84HH336 pKa = 4.87THH338 pKa = 6.58ASLLLNSGIPYY349 pKa = 10.0KK350 pKa = 10.34EE351 pKa = 3.8LQHH354 pKa = 6.73RR355 pKa = 11.84LGHH358 pKa = 5.38SRR360 pKa = 11.84ISITMDD366 pKa = 2.92IYY368 pKa = 11.54SHH370 pKa = 7.04LSKK373 pKa = 10.96EE374 pKa = 4.11NAKK377 pKa = 10.48NAVAFYY383 pKa = 10.59EE384 pKa = 4.29KK385 pKa = 10.97ALGNLL390 pKa = 3.69
Molecular weight: 44.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3120 |
47 |
447 |
183.5 |
21.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.282 ± 0.453 | 0.577 ± 0.122 |
5.865 ± 0.612 | 8.013 ± 0.691 |
3.654 ± 0.326 | 4.327 ± 0.378 |
1.923 ± 0.427 | 7.115 ± 0.439 |
8.622 ± 0.716 | 10.192 ± 0.462 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.756 ± 0.383 | 6.026 ± 0.582 |
2.821 ± 0.173 | 4.712 ± 0.43 |
5.385 ± 0.396 | 5.096 ± 0.571 |
6.41 ± 0.527 | 4.551 ± 0.274 |
0.865 ± 0.151 | 4.808 ± 0.34 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
