
Trypanosoma rangeli
Taxonomy: cellular organisms; Eukaryota; Discoba; Euglenozoa; Kinetoplastea; Metakinetoplastina; Trypanosomatida; Trypanosomatidae; Trypanosoma; Herpetosoma
Average proteome isoelectric point is 6.86
Get precalculated fractions of proteins
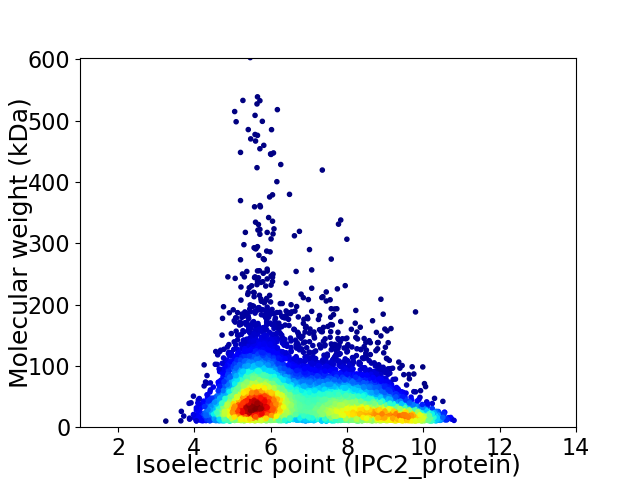
Virtual 2D-PAGE plot for 10070 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A422NHB0|A0A422NHB0_TRYRA Uncharacterized protein OS=Trypanosoma rangeli OX=5698 GN=TraAM80_04960 PE=4 SV=1
MM1 pKa = 7.18FAVDD5 pKa = 5.44LLDD8 pKa = 4.35ALAEE12 pKa = 4.01AAVILLFDD20 pKa = 4.1WTPTSACGDD29 pKa = 3.69EE30 pKa = 5.45DD31 pKa = 4.34NDD33 pKa = 4.43GEE35 pKa = 4.32IGSEE39 pKa = 3.93RR40 pKa = 11.84FTGLNSWSSLDD51 pKa = 3.38SCFPRR56 pKa = 11.84SQMNTTLSANNASLTASRR74 pKa = 11.84TLSLDD79 pKa = 3.16CSSFLQVAAEE89 pKa = 4.75EE90 pKa = 4.46IQTTPSEE97 pKa = 4.17QLVGSQDD104 pKa = 3.45HH105 pKa = 5.82YY106 pKa = 11.38CWTNEE111 pKa = 3.57YY112 pKa = 10.3EE113 pKa = 4.43YY114 pKa = 10.76FNSSSSTPSSWAVPVRR130 pKa = 11.84SLEE133 pKa = 4.04QKK135 pKa = 10.44
MM1 pKa = 7.18FAVDD5 pKa = 5.44LLDD8 pKa = 4.35ALAEE12 pKa = 4.01AAVILLFDD20 pKa = 4.1WTPTSACGDD29 pKa = 3.69EE30 pKa = 5.45DD31 pKa = 4.34NDD33 pKa = 4.43GEE35 pKa = 4.32IGSEE39 pKa = 3.93RR40 pKa = 11.84FTGLNSWSSLDD51 pKa = 3.38SCFPRR56 pKa = 11.84SQMNTTLSANNASLTASRR74 pKa = 11.84TLSLDD79 pKa = 3.16CSSFLQVAAEE89 pKa = 4.75EE90 pKa = 4.46IQTTPSEE97 pKa = 4.17QLVGSQDD104 pKa = 3.45HH105 pKa = 5.82YY106 pKa = 11.38CWTNEE111 pKa = 3.57YY112 pKa = 10.3EE113 pKa = 4.43YY114 pKa = 10.76FNSSSSTPSSWAVPVRR130 pKa = 11.84SLEE133 pKa = 4.04QKK135 pKa = 10.44
Molecular weight: 14.77 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A422NW00|A0A422NW00_TRYRA V-type proton ATPase proteolipid subunit OS=Trypanosoma rangeli OX=5698 GN=TraAM80_02012 PE=3 SV=1
MM1 pKa = 7.77PSTVARR7 pKa = 11.84EE8 pKa = 3.91RR9 pKa = 11.84NKK11 pKa = 10.56AAVRR15 pKa = 11.84RR16 pKa = 11.84LFARR20 pKa = 11.84DD21 pKa = 3.87HH22 pKa = 6.42VPSPAPWKK30 pKa = 10.4CHH32 pKa = 5.01TVHH35 pKa = 6.73CADD38 pKa = 3.42GTLPPGMHH46 pKa = 7.24LSKK49 pKa = 10.56QCEE52 pKa = 3.97LAHH55 pKa = 5.37EE56 pKa = 4.61VGRR59 pKa = 11.84GTAAAVAVAAAAATSSPHH77 pKa = 5.74SRR79 pKa = 11.84AVDD82 pKa = 3.47YY83 pKa = 10.96GNPFCSCYY91 pKa = 10.96SMNRR95 pKa = 11.84KK96 pKa = 8.53IARR99 pKa = 11.84HH100 pKa = 4.77EE101 pKa = 4.05WATSRR106 pKa = 11.84AEE108 pKa = 3.93KK109 pKa = 10.65GKK111 pKa = 10.09LQGGPLNPHH120 pKa = 5.91KK121 pKa = 10.45SARR124 pKa = 11.84EE125 pKa = 3.29RR126 pKa = 11.84RR127 pKa = 11.84AAIEE131 pKa = 3.94HH132 pKa = 6.52EE133 pKa = 4.28EE134 pKa = 4.21AILRR138 pKa = 11.84YY139 pKa = 8.86EE140 pKa = 3.81LSIPRR145 pKa = 11.84RR146 pKa = 11.84RR147 pKa = 11.84VAGHH151 pKa = 7.44LLHH154 pKa = 6.78LTAAQLSTSQPMPRR168 pKa = 11.84ARR170 pKa = 11.84TMDD173 pKa = 3.48RR174 pKa = 11.84LVEE177 pKa = 3.98HH178 pKa = 7.24RR179 pKa = 11.84KK180 pKa = 10.04GCWVLHH186 pKa = 5.23NPYY189 pKa = 9.85RR190 pKa = 11.84QQLLRR195 pKa = 11.84GEE197 pKa = 4.25PVMTRR202 pKa = 11.84IMHH205 pKa = 5.98GEE207 pKa = 3.97FYY209 pKa = 10.61EE210 pKa = 4.18PQRR213 pKa = 11.84NEE215 pKa = 3.45QNTINFSQANKK226 pKa = 9.79RR227 pKa = 11.84HH228 pKa = 4.85VQVASSINHH237 pKa = 6.13AMHH240 pKa = 6.98SSRR243 pKa = 11.84NTRR246 pKa = 3.1
MM1 pKa = 7.77PSTVARR7 pKa = 11.84EE8 pKa = 3.91RR9 pKa = 11.84NKK11 pKa = 10.56AAVRR15 pKa = 11.84RR16 pKa = 11.84LFARR20 pKa = 11.84DD21 pKa = 3.87HH22 pKa = 6.42VPSPAPWKK30 pKa = 10.4CHH32 pKa = 5.01TVHH35 pKa = 6.73CADD38 pKa = 3.42GTLPPGMHH46 pKa = 7.24LSKK49 pKa = 10.56QCEE52 pKa = 3.97LAHH55 pKa = 5.37EE56 pKa = 4.61VGRR59 pKa = 11.84GTAAAVAVAAAAATSSPHH77 pKa = 5.74SRR79 pKa = 11.84AVDD82 pKa = 3.47YY83 pKa = 10.96GNPFCSCYY91 pKa = 10.96SMNRR95 pKa = 11.84KK96 pKa = 8.53IARR99 pKa = 11.84HH100 pKa = 4.77EE101 pKa = 4.05WATSRR106 pKa = 11.84AEE108 pKa = 3.93KK109 pKa = 10.65GKK111 pKa = 10.09LQGGPLNPHH120 pKa = 5.91KK121 pKa = 10.45SARR124 pKa = 11.84EE125 pKa = 3.29RR126 pKa = 11.84RR127 pKa = 11.84AAIEE131 pKa = 3.94HH132 pKa = 6.52EE133 pKa = 4.28EE134 pKa = 4.21AILRR138 pKa = 11.84YY139 pKa = 8.86EE140 pKa = 3.81LSIPRR145 pKa = 11.84RR146 pKa = 11.84RR147 pKa = 11.84VAGHH151 pKa = 7.44LLHH154 pKa = 6.78LTAAQLSTSQPMPRR168 pKa = 11.84ARR170 pKa = 11.84TMDD173 pKa = 3.48RR174 pKa = 11.84LVEE177 pKa = 3.98HH178 pKa = 7.24RR179 pKa = 11.84KK180 pKa = 10.04GCWVLHH186 pKa = 5.23NPYY189 pKa = 9.85RR190 pKa = 11.84QQLLRR195 pKa = 11.84GEE197 pKa = 4.25PVMTRR202 pKa = 11.84IMHH205 pKa = 5.98GEE207 pKa = 3.97FYY209 pKa = 10.61EE210 pKa = 4.18PQRR213 pKa = 11.84NEE215 pKa = 3.45QNTINFSQANKK226 pKa = 9.79RR227 pKa = 11.84HH228 pKa = 4.85VQVASSINHH237 pKa = 6.13AMHH240 pKa = 6.98SSRR243 pKa = 11.84NTRR246 pKa = 3.1
Molecular weight: 27.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4520539 |
93 |
5642 |
448.9 |
49.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.199 ± 0.026 | 2.138 ± 0.012 |
4.771 ± 0.015 | 6.859 ± 0.024 |
3.636 ± 0.017 | 6.549 ± 0.02 |
2.672 ± 0.011 | 3.6 ± 0.017 |
4.123 ± 0.021 | 9.986 ± 0.024 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.471 ± 0.009 | 3.223 ± 0.013 |
4.995 ± 0.019 | 3.996 ± 0.016 |
7.276 ± 0.024 | 7.526 ± 0.024 |
5.691 ± 0.017 | 7.577 ± 0.018 |
1.225 ± 0.008 | 2.427 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
