
Poecilia reticulata (Guppy) (Acanthophacelus reticulatus)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii; Actinopteri; Neopterygii; Teleostei; Osteoglossocephalai; Clupeocephala; Euteleosteomorpha; Neoteleostei;
Average proteome isoelectric point is 6.64
Get precalculated fractions of proteins
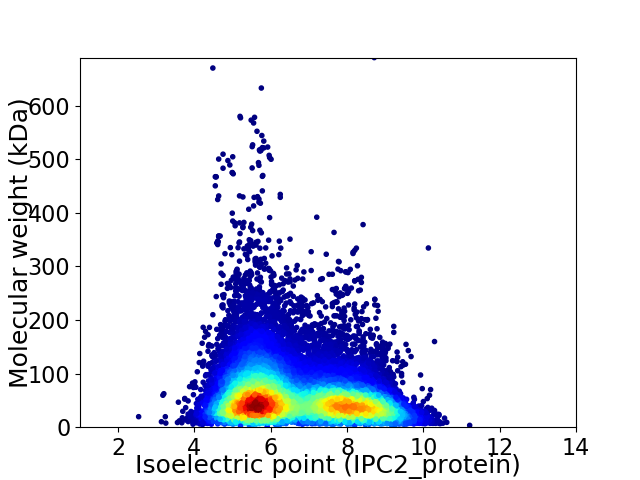
Virtual 2D-PAGE plot for 33944 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3P9MV63|A0A3P9MV63_POERE Nectin-4-like OS=Poecilia reticulata OX=8081 PE=4 SV=1
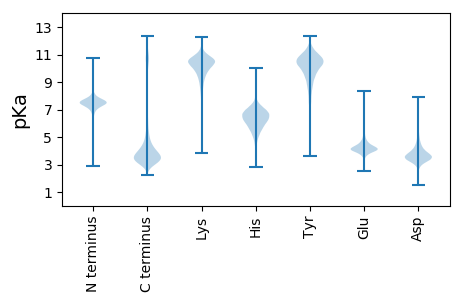
FF1 pKa = 7.41LKK3 pKa = 9.55NTVMEE8 pKa = 4.7CEE10 pKa = 3.94ACGMGGIQPVSPCEE24 pKa = 4.04PNPCHH29 pKa = 6.51PGVRR33 pKa = 11.84CSEE36 pKa = 4.3SPEE39 pKa = 4.23GPQCGACPDD48 pKa = 3.54GMEE51 pKa = 4.81GNGTRR56 pKa = 11.84CTDD59 pKa = 3.04VDD61 pKa = 3.71EE62 pKa = 5.13CKK64 pKa = 10.89VMPCHH69 pKa = 6.36MGVRR73 pKa = 11.84CINTSPGFRR82 pKa = 11.84CGSCPAGYY90 pKa = 8.41TGPQVQGVGLAYY102 pKa = 9.21ATANKK107 pKa = 9.14QVCRR111 pKa = 11.84DD112 pKa = 3.05IDD114 pKa = 3.69EE115 pKa = 5.41CEE117 pKa = 4.05GSSNGGCVEE126 pKa = 4.67NSVCLNTPGSFRR138 pKa = 11.84CGPCKK143 pKa = 9.85PGYY146 pKa = 10.65VGDD149 pKa = 3.78QRR151 pKa = 11.84LGCRR155 pKa = 11.84PEE157 pKa = 4.03RR158 pKa = 11.84TCGNGQPNPCHH169 pKa = 6.72ASADD173 pKa = 4.22CIVHH177 pKa = 6.53RR178 pKa = 11.84EE179 pKa = 4.11GTIEE183 pKa = 4.18CQCGVGWAGNGYY195 pKa = 9.9LCATDD200 pKa = 3.48TDD202 pKa = 3.7IDD204 pKa = 3.98GFPDD208 pKa = 3.5EE209 pKa = 5.98RR210 pKa = 11.84LDD212 pKa = 5.83CPDD215 pKa = 3.73LTCNKK220 pKa = 9.57DD221 pKa = 3.22NCLTVPNSGQEE232 pKa = 3.99DD233 pKa = 4.13ADD235 pKa = 3.65NDD237 pKa = 5.1GIGDD241 pKa = 4.07ACDD244 pKa = 4.46DD245 pKa = 4.56DD246 pKa = 6.96ADD248 pKa = 4.58GDD250 pKa = 4.67GIPNTQDD257 pKa = 2.74NCKK260 pKa = 9.99LVPNVDD266 pKa = 3.27QTNIDD271 pKa = 3.44EE272 pKa = 5.46DD273 pKa = 4.52DD274 pKa = 4.85FGDD277 pKa = 4.0ACDD280 pKa = 3.29NCRR283 pKa = 11.84AVKK286 pKa = 10.88NNDD289 pKa = 3.36QKK291 pKa = 11.3DD292 pKa = 3.45TDD294 pKa = 4.1LDD296 pKa = 3.97KK297 pKa = 11.72LGDD300 pKa = 3.86EE301 pKa = 5.26CDD303 pKa = 3.48EE304 pKa = 5.27DD305 pKa = 3.85MDD307 pKa = 5.78GDD309 pKa = 4.5GIINVMDD316 pKa = 3.41NCKK319 pKa = 9.88RR320 pKa = 11.84VPNVDD325 pKa = 3.03QKK327 pKa = 11.72DD328 pKa = 3.56RR329 pKa = 11.84DD330 pKa = 3.58GDD332 pKa = 4.12KK333 pKa = 11.46VGDD336 pKa = 4.05ACDD339 pKa = 3.38SCPYY343 pKa = 10.12VPNPDD348 pKa = 4.61QMDD351 pKa = 4.03ADD353 pKa = 4.07NDD355 pKa = 4.82LIGDD359 pKa = 3.97PCDD362 pKa = 3.62TNKK365 pKa = 10.93DD366 pKa = 3.49SDD368 pKa = 4.16GDD370 pKa = 3.81GHH372 pKa = 6.91QDD374 pKa = 4.2SRR376 pKa = 11.84DD377 pKa = 3.47NCPAVINSSQLDD389 pKa = 3.5TDD391 pKa = 3.78KK392 pKa = 11.37DD393 pKa = 4.07GKK395 pKa = 11.12GDD397 pKa = 3.71EE398 pKa = 5.18CDD400 pKa = 5.95DD401 pKa = 5.62DD402 pKa = 7.29DD403 pKa = 7.55DD404 pKa = 7.26DD405 pKa = 7.31DD406 pKa = 6.25GIPDD410 pKa = 5.2LLPPGPDD417 pKa = 2.92NCRR420 pKa = 11.84LVPNPLQEE428 pKa = 5.34DD429 pKa = 4.06LDD431 pKa = 5.15GDD433 pKa = 4.37GVGDD437 pKa = 3.69VCEE440 pKa = 4.78KK441 pKa = 11.14DD442 pKa = 3.38FDD444 pKa = 4.17NDD446 pKa = 4.31TIIDD450 pKa = 5.0PIDD453 pKa = 3.84ACPEE457 pKa = 3.91NAEE460 pKa = 4.1VTLTDD465 pKa = 3.59FRR467 pKa = 11.84EE468 pKa = 4.27YY469 pKa = 9.18QTVVLDD475 pKa = 4.15PEE477 pKa = 5.32GDD479 pKa = 3.65AQIDD483 pKa = 4.13PNWVVLNQGRR493 pKa = 11.84EE494 pKa = 3.9IVQTMNSDD502 pKa = 3.19PGLAVGYY509 pKa = 7.12TAFSGVDD516 pKa = 3.72FEE518 pKa = 5.24GTFHH522 pKa = 6.81VNTVTDD528 pKa = 3.57DD529 pKa = 4.08DD530 pKa = 4.35YY531 pKa = 12.0AGFIFGYY538 pKa = 9.58QDD540 pKa = 2.83SSSFYY545 pKa = 9.91VVMWKK550 pKa = 9.52QVEE553 pKa = 4.18QIYY556 pKa = 8.33WQANPFRR563 pKa = 11.84AVAQQGIQLKK573 pKa = 9.81AVKK576 pKa = 10.16SSTGPGEE583 pKa = 4.16NLRR586 pKa = 11.84NALWHH591 pKa = 6.37TGDD594 pKa = 3.91TGDD597 pKa = 4.85QVKK600 pKa = 10.34LLWKK604 pKa = 10.29DD605 pKa = 3.3PRR607 pKa = 11.84NVGWKK612 pKa = 10.34DD613 pKa = 3.02KK614 pKa = 10.83TSYY617 pKa = 10.52RR618 pKa = 11.84WFLQHH623 pKa = 6.87RR624 pKa = 11.84PADD627 pKa = 3.73GYY629 pKa = 10.45IRR631 pKa = 11.84VRR633 pKa = 11.84FYY635 pKa = 11.04EE636 pKa = 4.86GPQMVADD643 pKa = 3.78TGVIIDD649 pKa = 3.35ATMRR653 pKa = 11.84GGRR656 pKa = 11.84LGVFCFSQEE665 pKa = 3.95NIIWANLRR673 pKa = 11.84YY674 pKa = 9.71RR675 pKa = 11.84CNDD678 pKa = 3.51TLPGDD683 pKa = 3.68FDD685 pKa = 3.85TYY687 pKa = 10.14RR688 pKa = 11.84TQQVQLVAA696 pKa = 4.41
FF1 pKa = 7.41LKK3 pKa = 9.55NTVMEE8 pKa = 4.7CEE10 pKa = 3.94ACGMGGIQPVSPCEE24 pKa = 4.04PNPCHH29 pKa = 6.51PGVRR33 pKa = 11.84CSEE36 pKa = 4.3SPEE39 pKa = 4.23GPQCGACPDD48 pKa = 3.54GMEE51 pKa = 4.81GNGTRR56 pKa = 11.84CTDD59 pKa = 3.04VDD61 pKa = 3.71EE62 pKa = 5.13CKK64 pKa = 10.89VMPCHH69 pKa = 6.36MGVRR73 pKa = 11.84CINTSPGFRR82 pKa = 11.84CGSCPAGYY90 pKa = 8.41TGPQVQGVGLAYY102 pKa = 9.21ATANKK107 pKa = 9.14QVCRR111 pKa = 11.84DD112 pKa = 3.05IDD114 pKa = 3.69EE115 pKa = 5.41CEE117 pKa = 4.05GSSNGGCVEE126 pKa = 4.67NSVCLNTPGSFRR138 pKa = 11.84CGPCKK143 pKa = 9.85PGYY146 pKa = 10.65VGDD149 pKa = 3.78QRR151 pKa = 11.84LGCRR155 pKa = 11.84PEE157 pKa = 4.03RR158 pKa = 11.84TCGNGQPNPCHH169 pKa = 6.72ASADD173 pKa = 4.22CIVHH177 pKa = 6.53RR178 pKa = 11.84EE179 pKa = 4.11GTIEE183 pKa = 4.18CQCGVGWAGNGYY195 pKa = 9.9LCATDD200 pKa = 3.48TDD202 pKa = 3.7IDD204 pKa = 3.98GFPDD208 pKa = 3.5EE209 pKa = 5.98RR210 pKa = 11.84LDD212 pKa = 5.83CPDD215 pKa = 3.73LTCNKK220 pKa = 9.57DD221 pKa = 3.22NCLTVPNSGQEE232 pKa = 3.99DD233 pKa = 4.13ADD235 pKa = 3.65NDD237 pKa = 5.1GIGDD241 pKa = 4.07ACDD244 pKa = 4.46DD245 pKa = 4.56DD246 pKa = 6.96ADD248 pKa = 4.58GDD250 pKa = 4.67GIPNTQDD257 pKa = 2.74NCKK260 pKa = 9.99LVPNVDD266 pKa = 3.27QTNIDD271 pKa = 3.44EE272 pKa = 5.46DD273 pKa = 4.52DD274 pKa = 4.85FGDD277 pKa = 4.0ACDD280 pKa = 3.29NCRR283 pKa = 11.84AVKK286 pKa = 10.88NNDD289 pKa = 3.36QKK291 pKa = 11.3DD292 pKa = 3.45TDD294 pKa = 4.1LDD296 pKa = 3.97KK297 pKa = 11.72LGDD300 pKa = 3.86EE301 pKa = 5.26CDD303 pKa = 3.48EE304 pKa = 5.27DD305 pKa = 3.85MDD307 pKa = 5.78GDD309 pKa = 4.5GIINVMDD316 pKa = 3.41NCKK319 pKa = 9.88RR320 pKa = 11.84VPNVDD325 pKa = 3.03QKK327 pKa = 11.72DD328 pKa = 3.56RR329 pKa = 11.84DD330 pKa = 3.58GDD332 pKa = 4.12KK333 pKa = 11.46VGDD336 pKa = 4.05ACDD339 pKa = 3.38SCPYY343 pKa = 10.12VPNPDD348 pKa = 4.61QMDD351 pKa = 4.03ADD353 pKa = 4.07NDD355 pKa = 4.82LIGDD359 pKa = 3.97PCDD362 pKa = 3.62TNKK365 pKa = 10.93DD366 pKa = 3.49SDD368 pKa = 4.16GDD370 pKa = 3.81GHH372 pKa = 6.91QDD374 pKa = 4.2SRR376 pKa = 11.84DD377 pKa = 3.47NCPAVINSSQLDD389 pKa = 3.5TDD391 pKa = 3.78KK392 pKa = 11.37DD393 pKa = 4.07GKK395 pKa = 11.12GDD397 pKa = 3.71EE398 pKa = 5.18CDD400 pKa = 5.95DD401 pKa = 5.62DD402 pKa = 7.29DD403 pKa = 7.55DD404 pKa = 7.26DD405 pKa = 7.31DD406 pKa = 6.25GIPDD410 pKa = 5.2LLPPGPDD417 pKa = 2.92NCRR420 pKa = 11.84LVPNPLQEE428 pKa = 5.34DD429 pKa = 4.06LDD431 pKa = 5.15GDD433 pKa = 4.37GVGDD437 pKa = 3.69VCEE440 pKa = 4.78KK441 pKa = 11.14DD442 pKa = 3.38FDD444 pKa = 4.17NDD446 pKa = 4.31TIIDD450 pKa = 5.0PIDD453 pKa = 3.84ACPEE457 pKa = 3.91NAEE460 pKa = 4.1VTLTDD465 pKa = 3.59FRR467 pKa = 11.84EE468 pKa = 4.27YY469 pKa = 9.18QTVVLDD475 pKa = 4.15PEE477 pKa = 5.32GDD479 pKa = 3.65AQIDD483 pKa = 4.13PNWVVLNQGRR493 pKa = 11.84EE494 pKa = 3.9IVQTMNSDD502 pKa = 3.19PGLAVGYY509 pKa = 7.12TAFSGVDD516 pKa = 3.72FEE518 pKa = 5.24GTFHH522 pKa = 6.81VNTVTDD528 pKa = 3.57DD529 pKa = 4.08DD530 pKa = 4.35YY531 pKa = 12.0AGFIFGYY538 pKa = 9.58QDD540 pKa = 2.83SSSFYY545 pKa = 9.91VVMWKK550 pKa = 9.52QVEE553 pKa = 4.18QIYY556 pKa = 8.33WQANPFRR563 pKa = 11.84AVAQQGIQLKK573 pKa = 9.81AVKK576 pKa = 10.16SSTGPGEE583 pKa = 4.16NLRR586 pKa = 11.84NALWHH591 pKa = 6.37TGDD594 pKa = 3.91TGDD597 pKa = 4.85QVKK600 pKa = 10.34LLWKK604 pKa = 10.29DD605 pKa = 3.3PRR607 pKa = 11.84NVGWKK612 pKa = 10.34DD613 pKa = 3.02KK614 pKa = 10.83TSYY617 pKa = 10.52RR618 pKa = 11.84WFLQHH623 pKa = 6.87RR624 pKa = 11.84PADD627 pKa = 3.73GYY629 pKa = 10.45IRR631 pKa = 11.84VRR633 pKa = 11.84FYY635 pKa = 11.04EE636 pKa = 4.86GPQMVADD643 pKa = 3.78TGVIIDD649 pKa = 3.35ATMRR653 pKa = 11.84GGRR656 pKa = 11.84LGVFCFSQEE665 pKa = 3.95NIIWANLRR673 pKa = 11.84YY674 pKa = 9.71RR675 pKa = 11.84CNDD678 pKa = 3.51TLPGDD683 pKa = 3.68FDD685 pKa = 3.85TYY687 pKa = 10.14RR688 pKa = 11.84TQQVQLVAA696 pKa = 4.41
Molecular weight: 75.62 kDa
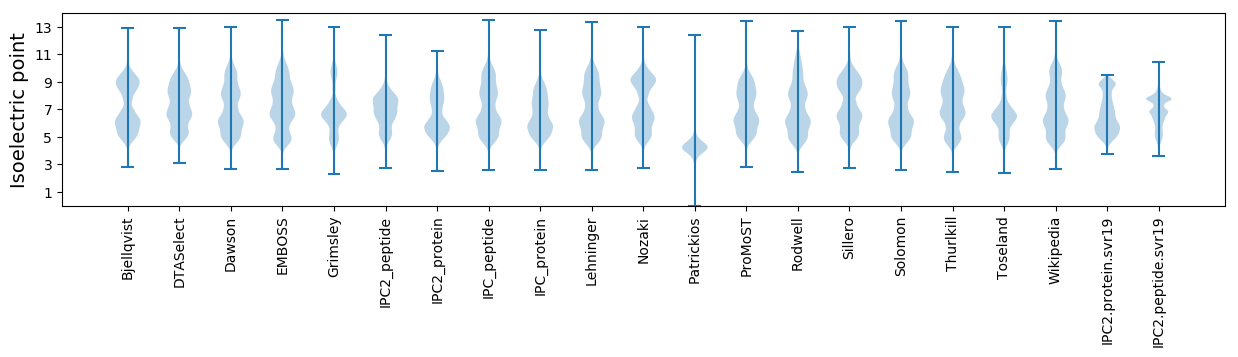
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3P9PAF3|A0A3P9PAF3_POERE Uncharacterized protein OS=Poecilia reticulata OX=8081 PE=4 SV=1
MM1 pKa = 7.1RR2 pKa = 11.84AKK4 pKa = 9.12WRR6 pKa = 11.84KK7 pKa = 9.1KK8 pKa = 9.32RR9 pKa = 11.84MRR11 pKa = 11.84RR12 pKa = 11.84LKK14 pKa = 10.08RR15 pKa = 11.84KK16 pKa = 8.21RR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 8.46MRR21 pKa = 11.84QRR23 pKa = 11.84SKK25 pKa = 11.41
MM1 pKa = 7.1RR2 pKa = 11.84AKK4 pKa = 9.12WRR6 pKa = 11.84KK7 pKa = 9.1KK8 pKa = 9.32RR9 pKa = 11.84MRR11 pKa = 11.84RR12 pKa = 11.84LKK14 pKa = 10.08RR15 pKa = 11.84KK16 pKa = 8.21RR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 8.46MRR21 pKa = 11.84QRR23 pKa = 11.84SKK25 pKa = 11.41
Molecular weight: 3.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
19349097 |
9 |
6188 |
570.0 |
63.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.505 ± 0.011 | 2.274 ± 0.011 |
5.185 ± 0.008 | 6.774 ± 0.016 |
3.796 ± 0.009 | 6.309 ± 0.016 |
2.615 ± 0.007 | 4.395 ± 0.009 |
5.685 ± 0.014 | 9.606 ± 0.018 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.34 ± 0.005 | 3.888 ± 0.008 |
5.669 ± 0.016 | 4.699 ± 0.011 |
5.661 ± 0.011 | 8.774 ± 0.017 |
5.523 ± 0.008 | 6.319 ± 0.011 |
1.161 ± 0.004 | 2.721 ± 0.007 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
