
Pseudoflavonifractor capillosus ATCC 29799
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Oscillospiraceae; Pseudoflavonifractor; Pseudoflavonifractor capillosus
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
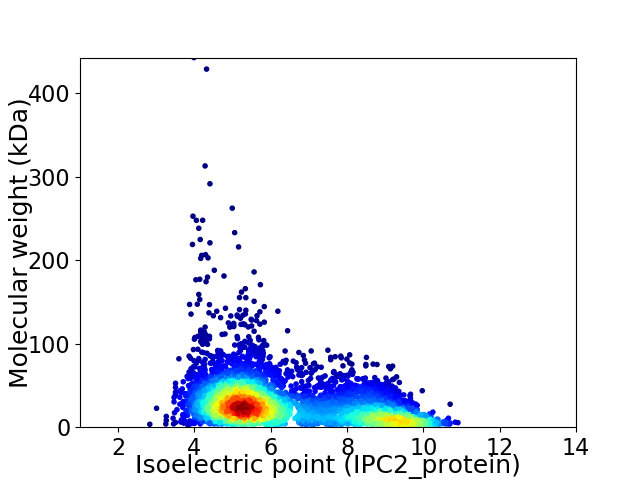
Virtual 2D-PAGE plot for 4804 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
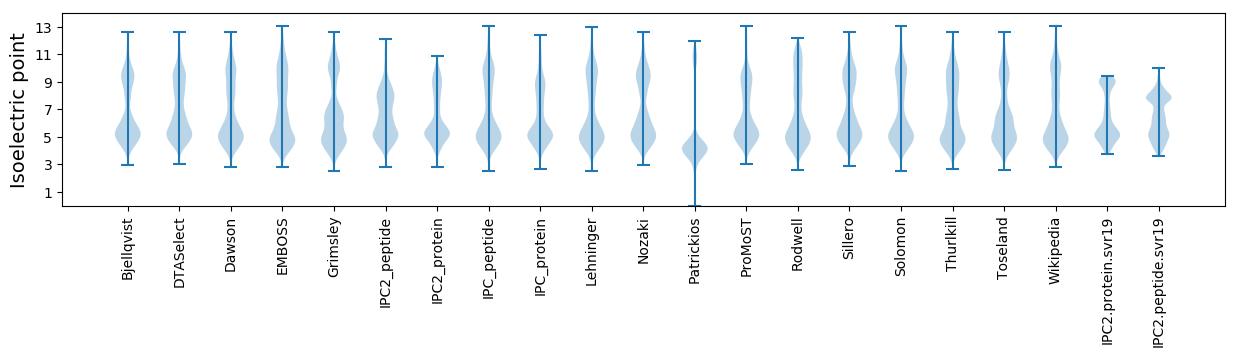
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A6NRS0|A6NRS0_9FIRM Uncharacterized protein OS=Pseudoflavonifractor capillosus ATCC 29799 OX=411467 GN=BACCAP_00898 PE=4 SV=1
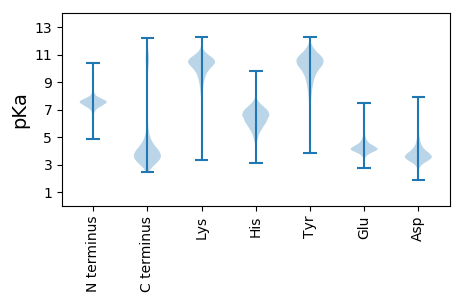
MM1 pKa = 7.48KK2 pKa = 10.13GKK4 pKa = 10.5KK5 pKa = 9.17IFSAALSLSLAAVLCLSGCTSAAAEE30 pKa = 4.38DD31 pKa = 4.57SNASSAVMPSVPPAEE46 pKa = 4.33TTVDD50 pKa = 3.71VPTSDD55 pKa = 3.79PLAPPEE61 pKa = 4.39GPVYY65 pKa = 10.47TIAQQHH71 pKa = 5.24QPDD74 pKa = 4.08LVRR77 pKa = 11.84VQGTVSVDD85 pKa = 2.88GDD87 pKa = 3.92RR88 pKa = 11.84ILVTNSNEE96 pKa = 3.38WAQYY100 pKa = 9.59PEE102 pKa = 4.15IVLSLEE108 pKa = 3.86KK109 pKa = 10.28DD110 pKa = 3.66TLILNAVDD118 pKa = 4.63GSAMAAEE125 pKa = 4.7DD126 pKa = 4.01LRR128 pKa = 11.84EE129 pKa = 4.11GEE131 pKa = 4.08YY132 pKa = 10.84VYY134 pKa = 10.83AYY136 pKa = 9.89VSPAMTDD143 pKa = 3.18SLPPITTAAVILAAIPADD161 pKa = 3.79YY162 pKa = 10.61AVPAYY167 pKa = 10.71SIVEE171 pKa = 3.95QVNVWEE177 pKa = 4.74GRR179 pKa = 11.84TSILMSDD186 pKa = 3.74DD187 pKa = 3.15VTYY190 pKa = 11.55SMSDD194 pKa = 3.17AEE196 pKa = 4.62LLAGPGFTCGTITLADD212 pKa = 3.56ITPGCGLLAWYY223 pKa = 10.29DD224 pKa = 3.61MLAEE228 pKa = 4.44SFPAQAWPVKK238 pKa = 10.34VMAIPSPYY246 pKa = 9.93DD247 pKa = 2.77AWAEE251 pKa = 4.02LEE253 pKa = 4.1PGRR256 pKa = 11.84LAVNGTEE263 pKa = 4.21VEE265 pKa = 4.24LTEE268 pKa = 5.07AEE270 pKa = 4.07QPYY273 pKa = 10.07VEE275 pKa = 5.48GEE277 pKa = 4.15TLMVPVRR284 pKa = 11.84AFAEE288 pKa = 4.02ALGCEE293 pKa = 4.78VIWNADD299 pKa = 3.4QPEE302 pKa = 4.67SISVWGEE309 pKa = 3.35GAEE312 pKa = 4.05YY313 pKa = 10.94LLTLGSDD320 pKa = 3.64RR321 pKa = 11.84AATGDD326 pKa = 3.79GGDD329 pKa = 3.7GTALTGAVTARR340 pKa = 11.84NGVSFLPADD349 pKa = 4.42DD350 pKa = 5.37LLNLHH355 pKa = 6.08NVRR358 pKa = 11.84AEE360 pKa = 4.15GLWPVV365 pKa = 3.43
MM1 pKa = 7.48KK2 pKa = 10.13GKK4 pKa = 10.5KK5 pKa = 9.17IFSAALSLSLAAVLCLSGCTSAAAEE30 pKa = 4.38DD31 pKa = 4.57SNASSAVMPSVPPAEE46 pKa = 4.33TTVDD50 pKa = 3.71VPTSDD55 pKa = 3.79PLAPPEE61 pKa = 4.39GPVYY65 pKa = 10.47TIAQQHH71 pKa = 5.24QPDD74 pKa = 4.08LVRR77 pKa = 11.84VQGTVSVDD85 pKa = 2.88GDD87 pKa = 3.92RR88 pKa = 11.84ILVTNSNEE96 pKa = 3.38WAQYY100 pKa = 9.59PEE102 pKa = 4.15IVLSLEE108 pKa = 3.86KK109 pKa = 10.28DD110 pKa = 3.66TLILNAVDD118 pKa = 4.63GSAMAAEE125 pKa = 4.7DD126 pKa = 4.01LRR128 pKa = 11.84EE129 pKa = 4.11GEE131 pKa = 4.08YY132 pKa = 10.84VYY134 pKa = 10.83AYY136 pKa = 9.89VSPAMTDD143 pKa = 3.18SLPPITTAAVILAAIPADD161 pKa = 3.79YY162 pKa = 10.61AVPAYY167 pKa = 10.71SIVEE171 pKa = 3.95QVNVWEE177 pKa = 4.74GRR179 pKa = 11.84TSILMSDD186 pKa = 3.74DD187 pKa = 3.15VTYY190 pKa = 11.55SMSDD194 pKa = 3.17AEE196 pKa = 4.62LLAGPGFTCGTITLADD212 pKa = 3.56ITPGCGLLAWYY223 pKa = 10.29DD224 pKa = 3.61MLAEE228 pKa = 4.44SFPAQAWPVKK238 pKa = 10.34VMAIPSPYY246 pKa = 9.93DD247 pKa = 2.77AWAEE251 pKa = 4.02LEE253 pKa = 4.1PGRR256 pKa = 11.84LAVNGTEE263 pKa = 4.21VEE265 pKa = 4.24LTEE268 pKa = 5.07AEE270 pKa = 4.07QPYY273 pKa = 10.07VEE275 pKa = 5.48GEE277 pKa = 4.15TLMVPVRR284 pKa = 11.84AFAEE288 pKa = 4.02ALGCEE293 pKa = 4.78VIWNADD299 pKa = 3.4QPEE302 pKa = 4.67SISVWGEE309 pKa = 3.35GAEE312 pKa = 4.05YY313 pKa = 10.94LLTLGSDD320 pKa = 3.64RR321 pKa = 11.84AATGDD326 pKa = 3.79GGDD329 pKa = 3.7GTALTGAVTARR340 pKa = 11.84NGVSFLPADD349 pKa = 4.42DD350 pKa = 5.37LLNLHH355 pKa = 6.08NVRR358 pKa = 11.84AEE360 pKa = 4.15GLWPVV365 pKa = 3.43
Molecular weight: 38.38 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A6NST1|A6NST1_9FIRM Uncharacterized protein OS=Pseudoflavonifractor capillosus ATCC 29799 OX=411467 GN=BACCAP_01260 PE=4 SV=1
MM1 pKa = 7.57IFFGMINHH9 pKa = 6.64TPSFSFLAQILWSSRR24 pKa = 11.84QAARR28 pKa = 11.84RR29 pKa = 11.84ARR31 pKa = 11.84FLIFWHH37 pKa = 6.5PNIIAAQNRR46 pKa = 11.84KK47 pKa = 9.01AVNPP51 pKa = 4.06
MM1 pKa = 7.57IFFGMINHH9 pKa = 6.64TPSFSFLAQILWSSRR24 pKa = 11.84QAARR28 pKa = 11.84RR29 pKa = 11.84ARR31 pKa = 11.84FLIFWHH37 pKa = 6.5PNIIAAQNRR46 pKa = 11.84KK47 pKa = 9.01AVNPP51 pKa = 4.06
Molecular weight: 5.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1302578 |
17 |
4122 |
271.1 |
30.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.327 ± 0.045 | 1.649 ± 0.02 |
5.672 ± 0.03 | 6.816 ± 0.037 |
3.867 ± 0.027 | 7.82 ± 0.041 |
1.817 ± 0.016 | 5.403 ± 0.03 |
4.683 ± 0.04 | 9.885 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.801 ± 0.021 | 3.342 ± 0.028 |
4.265 ± 0.026 | 3.523 ± 0.025 |
5.677 ± 0.047 | 5.794 ± 0.032 |
5.762 ± 0.049 | 7.147 ± 0.034 |
1.152 ± 0.013 | 3.598 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
