
Homo sapiens (Human)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Sarcopterygii; Dipnotetrapodomorpha; Tetrapoda; Amniota; Mammalia; Theria; Eutheria; Boreoeutheria; Euarchontoglires; Primates;
Average proteome isoelectric point is 6.71
Get precalculated fractions of proteins
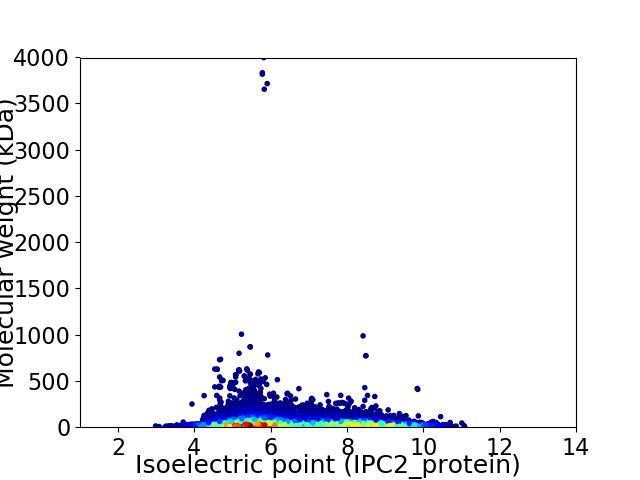
Virtual 2D-PAGE plot for 100100 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F2Z311|F2Z311_HUMAN Isoform of Q86TG1 Transmembrane protein 150A OS=Homo sapiens OX=9606 GN=TMEM150A PE=4 SV=1
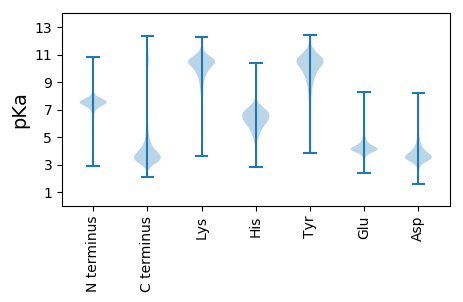
MM1 pKa = 7.5SEE3 pKa = 4.04KK4 pKa = 10.31KK5 pKa = 10.32QPVDD9 pKa = 3.98LGLLEE14 pKa = 4.72EE15 pKa = 5.08DD16 pKa = 4.95DD17 pKa = 4.2EE18 pKa = 5.16FEE20 pKa = 4.5EE21 pKa = 6.21FPAEE25 pKa = 3.92DD26 pKa = 3.4WAGLDD31 pKa = 3.67EE32 pKa = 6.8DD33 pKa = 4.65EE34 pKa = 5.59DD35 pKa = 3.98AHH37 pKa = 7.46VWEE40 pKa = 5.76DD41 pKa = 3.11NWDD44 pKa = 3.91DD45 pKa = 5.56DD46 pKa = 4.79NVEE49 pKa = 4.84DD50 pKa = 5.62DD51 pKa = 4.76FSNQLRR57 pKa = 11.84ATVLLMILVCEE68 pKa = 4.39TPYY71 pKa = 11.06GCYY74 pKa = 9.94VLHH77 pKa = 6.36QKK79 pKa = 10.5GRR81 pKa = 11.84MCSAFLCCC89 pKa = 6.08
MM1 pKa = 7.5SEE3 pKa = 4.04KK4 pKa = 10.31KK5 pKa = 10.32QPVDD9 pKa = 3.98LGLLEE14 pKa = 4.72EE15 pKa = 5.08DD16 pKa = 4.95DD17 pKa = 4.2EE18 pKa = 5.16FEE20 pKa = 4.5EE21 pKa = 6.21FPAEE25 pKa = 3.92DD26 pKa = 3.4WAGLDD31 pKa = 3.67EE32 pKa = 6.8DD33 pKa = 4.65EE34 pKa = 5.59DD35 pKa = 3.98AHH37 pKa = 7.46VWEE40 pKa = 5.76DD41 pKa = 3.11NWDD44 pKa = 3.91DD45 pKa = 5.56DD46 pKa = 4.79NVEE49 pKa = 4.84DD50 pKa = 5.62DD51 pKa = 4.76FSNQLRR57 pKa = 11.84ATVLLMILVCEE68 pKa = 4.39TPYY71 pKa = 11.06GCYY74 pKa = 9.94VLHH77 pKa = 6.36QKK79 pKa = 10.5GRR81 pKa = 11.84MCSAFLCCC89 pKa = 6.08
Molecular weight: 10.32 kDa
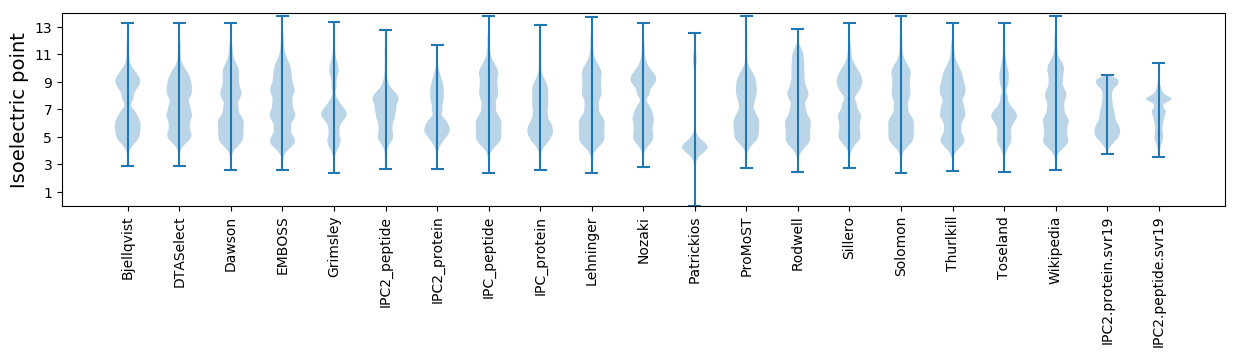
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q9UKM7|MA1B1_HUMAN Endoplasmic reticulum mannosyl-oligosaccharide 1 2-alpha-mannosidase OS=Homo sapiens OX=9606 GN=MAN1B1 PE=1 SV=2
MM1 pKa = 7.76RR2 pKa = 11.84RR3 pKa = 11.84PSTASLTRR11 pKa = 11.84TPSRR15 pKa = 11.84ASPTRR20 pKa = 11.84MPSRR24 pKa = 11.84ASLKK28 pKa = 7.11MTPFRR33 pKa = 11.84ASLTKK38 pKa = 9.87MEE40 pKa = 4.28STALLRR46 pKa = 11.84TLPRR50 pKa = 11.84ASLMRR55 pKa = 11.84TPTRR59 pKa = 11.84ASLMRR64 pKa = 11.84TPPRR68 pKa = 11.84ASPTRR73 pKa = 11.84KK74 pKa = 9.24PPRR77 pKa = 11.84ASPRR81 pKa = 11.84TPSRR85 pKa = 11.84ASPTRR90 pKa = 11.84RR91 pKa = 11.84LPRR94 pKa = 11.84ASPMGSPHH102 pKa = 6.87RR103 pKa = 11.84ASPMRR108 pKa = 11.84TPPRR112 pKa = 11.84ASPTGTPSTASPTGTPSSASPTGTPPRR139 pKa = 11.84ASPTGTPPRR148 pKa = 11.84AWATRR153 pKa = 11.84SPSTASLTRR162 pKa = 11.84TPSRR166 pKa = 11.84ASLTRR171 pKa = 11.84WPPRR175 pKa = 11.84ASPTRR180 pKa = 11.84TPPRR184 pKa = 11.84EE185 pKa = 4.12SPRR188 pKa = 11.84MSHH191 pKa = 6.34RR192 pKa = 11.84ASPTRR197 pKa = 11.84TPPRR201 pKa = 11.84ASPTRR206 pKa = 11.84RR207 pKa = 11.84PPRR210 pKa = 11.84ASPTRR215 pKa = 11.84TPPRR219 pKa = 11.84EE220 pKa = 4.02SLRR223 pKa = 11.84TSHH226 pKa = 6.8RR227 pKa = 11.84ASPTRR232 pKa = 11.84MPPRR236 pKa = 11.84ASPTRR241 pKa = 11.84RR242 pKa = 11.84PPRR245 pKa = 11.84ASPTGSPPRR254 pKa = 11.84ASPMTPPRR262 pKa = 11.84ASPRR266 pKa = 11.84TPPRR270 pKa = 11.84ASPTTTPSRR279 pKa = 11.84ASLTRR284 pKa = 11.84TPSWASPTTTPSRR297 pKa = 11.84ASLMKK302 pKa = 9.87MEE304 pKa = 4.46STVSITRR311 pKa = 11.84TPPRR315 pKa = 11.84ASPTGTPSRR324 pKa = 11.84ASPTGTPSRR333 pKa = 11.84ASLTGSPSRR342 pKa = 11.84ASLTGTPSRR351 pKa = 11.84ASLIGTPSRR360 pKa = 11.84ASLIGTPSRR369 pKa = 11.84ASLTGTPPRR378 pKa = 11.84ASLTGTSSTASLTRR392 pKa = 11.84TPSRR396 pKa = 11.84ASLTRR401 pKa = 11.84TQSSSSLTRR410 pKa = 11.84TPSMASLTRR419 pKa = 11.84TPPRR423 pKa = 11.84ASLTRR428 pKa = 11.84TPPRR432 pKa = 11.84ASLTRR437 pKa = 11.84TPPRR441 pKa = 11.84ASLTRR446 pKa = 11.84TPPRR450 pKa = 11.84ASLTRR455 pKa = 11.84TPSMVSLKK463 pKa = 10.16RR464 pKa = 11.84SPSRR468 pKa = 11.84ASLTRR473 pKa = 11.84TPSRR477 pKa = 11.84ASLTMTPSRR486 pKa = 11.84ASLTRR491 pKa = 11.84TPSTASLTGTPPTASLTRR509 pKa = 11.84TPPTASLTRR518 pKa = 11.84SPPTASLTRR527 pKa = 11.84TPSTASLTRR536 pKa = 11.84MPSTASLTRR545 pKa = 11.84KK546 pKa = 10.27SNVNQQCPASTPSSEE561 pKa = 4.48VISS564 pKa = 3.92
MM1 pKa = 7.76RR2 pKa = 11.84RR3 pKa = 11.84PSTASLTRR11 pKa = 11.84TPSRR15 pKa = 11.84ASPTRR20 pKa = 11.84MPSRR24 pKa = 11.84ASLKK28 pKa = 7.11MTPFRR33 pKa = 11.84ASLTKK38 pKa = 9.87MEE40 pKa = 4.28STALLRR46 pKa = 11.84TLPRR50 pKa = 11.84ASLMRR55 pKa = 11.84TPTRR59 pKa = 11.84ASLMRR64 pKa = 11.84TPPRR68 pKa = 11.84ASPTRR73 pKa = 11.84KK74 pKa = 9.24PPRR77 pKa = 11.84ASPRR81 pKa = 11.84TPSRR85 pKa = 11.84ASPTRR90 pKa = 11.84RR91 pKa = 11.84LPRR94 pKa = 11.84ASPMGSPHH102 pKa = 6.87RR103 pKa = 11.84ASPMRR108 pKa = 11.84TPPRR112 pKa = 11.84ASPTGTPSTASPTGTPSSASPTGTPPRR139 pKa = 11.84ASPTGTPPRR148 pKa = 11.84AWATRR153 pKa = 11.84SPSTASLTRR162 pKa = 11.84TPSRR166 pKa = 11.84ASLTRR171 pKa = 11.84WPPRR175 pKa = 11.84ASPTRR180 pKa = 11.84TPPRR184 pKa = 11.84EE185 pKa = 4.12SPRR188 pKa = 11.84MSHH191 pKa = 6.34RR192 pKa = 11.84ASPTRR197 pKa = 11.84TPPRR201 pKa = 11.84ASPTRR206 pKa = 11.84RR207 pKa = 11.84PPRR210 pKa = 11.84ASPTRR215 pKa = 11.84TPPRR219 pKa = 11.84EE220 pKa = 4.02SLRR223 pKa = 11.84TSHH226 pKa = 6.8RR227 pKa = 11.84ASPTRR232 pKa = 11.84MPPRR236 pKa = 11.84ASPTRR241 pKa = 11.84RR242 pKa = 11.84PPRR245 pKa = 11.84ASPTGSPPRR254 pKa = 11.84ASPMTPPRR262 pKa = 11.84ASPRR266 pKa = 11.84TPPRR270 pKa = 11.84ASPTTTPSRR279 pKa = 11.84ASLTRR284 pKa = 11.84TPSWASPTTTPSRR297 pKa = 11.84ASLMKK302 pKa = 9.87MEE304 pKa = 4.46STVSITRR311 pKa = 11.84TPPRR315 pKa = 11.84ASPTGTPSRR324 pKa = 11.84ASPTGTPSRR333 pKa = 11.84ASLTGSPSRR342 pKa = 11.84ASLTGTPSRR351 pKa = 11.84ASLIGTPSRR360 pKa = 11.84ASLIGTPSRR369 pKa = 11.84ASLTGTPPRR378 pKa = 11.84ASLTGTSSTASLTRR392 pKa = 11.84TPSRR396 pKa = 11.84ASLTRR401 pKa = 11.84TQSSSSLTRR410 pKa = 11.84TPSMASLTRR419 pKa = 11.84TPPRR423 pKa = 11.84ASLTRR428 pKa = 11.84TPPRR432 pKa = 11.84ASLTRR437 pKa = 11.84TPPRR441 pKa = 11.84ASLTRR446 pKa = 11.84TPPRR450 pKa = 11.84ASLTRR455 pKa = 11.84TPSMVSLKK463 pKa = 10.16RR464 pKa = 11.84SPSRR468 pKa = 11.84ASLTRR473 pKa = 11.84TPSRR477 pKa = 11.84ASLTMTPSRR486 pKa = 11.84ASLTRR491 pKa = 11.84TPSTASLTGTPPTASLTRR509 pKa = 11.84TPPTASLTRR518 pKa = 11.84SPPTASLTRR527 pKa = 11.84TPSTASLTRR536 pKa = 11.84MPSTASLTRR545 pKa = 11.84KK546 pKa = 10.27SNVNQQCPASTPSSEE561 pKa = 4.48VISS564 pKa = 3.92
Molecular weight: 59.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
40284240 |
2 |
35991 |
402.4 |
44.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.92 ± 0.009 | 2.167 ± 0.008 |
4.835 ± 0.006 | 7.207 ± 0.013 |
3.549 ± 0.007 | 6.503 ± 0.014 |
2.577 ± 0.006 | 4.346 ± 0.01 |
5.77 ± 0.015 | 9.837 ± 0.018 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.185 ± 0.005 | 3.605 ± 0.007 |
6.3 ± 0.013 | 4.81 ± 0.013 |
5.645 ± 0.01 | 8.408 ± 0.017 |
5.466 ± 0.021 | 6.019 ± 0.014 |
1.223 ± 0.003 | 2.607 ± 0.005 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
