
Haloquadratum sp. J07HQX50
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Haloferacales; Haloferacaceae; Haloquadratum; unclassified Haloquadratum
Average proteome isoelectric point is 5.42
Get precalculated fractions of proteins
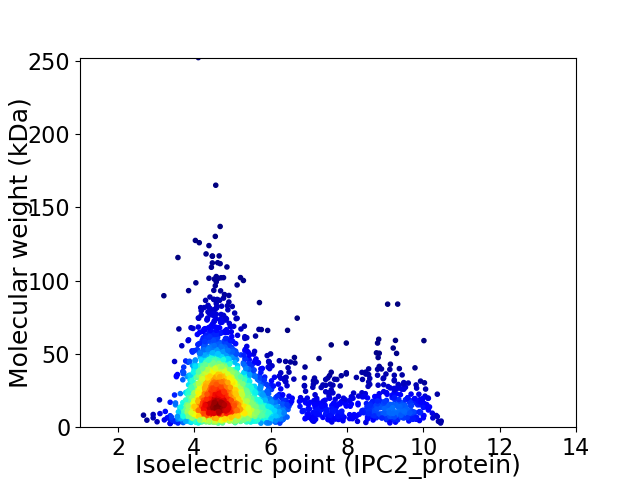
Virtual 2D-PAGE plot for 2865 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|U1PZW5|U1PZW5_9EURY Spermidine/putrescine-binding periplasmic protein OS=Haloquadratum sp. J07HQX50 OX=1238426 GN=J07HQX50_02174 PE=4 SV=1
MM1 pKa = 7.76TDD3 pKa = 3.61DD4 pKa = 4.65SSPSSDD10 pKa = 4.53EE11 pKa = 3.76EE12 pKa = 3.9LTYY15 pKa = 11.33AKK17 pKa = 10.54AGVDD21 pKa = 3.08IDD23 pKa = 4.05ASEE26 pKa = 4.3SATDD30 pKa = 3.84ALLQAVGEE38 pKa = 4.52TPGDD42 pKa = 3.58YY43 pKa = 10.89AGLIDD48 pKa = 5.7IGNQYY53 pKa = 10.86LAVTTDD59 pKa = 3.45GVGTKK64 pKa = 10.53LLIAEE69 pKa = 5.67AIAEE73 pKa = 4.14YY74 pKa = 9.31STIGIDD80 pKa = 3.56CVAMNVNDD88 pKa = 4.12LVAEE92 pKa = 4.65GVRR95 pKa = 11.84PIGFVDD101 pKa = 3.97YY102 pKa = 10.83LAVDD106 pKa = 3.94NPDD109 pKa = 3.4EE110 pKa = 5.85DD111 pKa = 3.52IASQIGVGLQSGADD125 pKa = 3.41RR126 pKa = 11.84ADD128 pKa = 2.91ISLIGGEE135 pKa = 4.16TAVMPEE141 pKa = 4.27VINGFDD147 pKa = 4.1LAGTCVGLAPKK158 pKa = 10.28SDD160 pKa = 3.64VLTQTAQPGDD170 pKa = 3.88IIAGFPSSGIHH181 pKa = 6.89SNGLTLARR189 pKa = 11.84KK190 pKa = 9.52AIQNQFSYY198 pKa = 10.26TDD200 pKa = 3.6QYY202 pKa = 11.6PEE204 pKa = 3.83SQYY207 pKa = 11.77NSIAEE212 pKa = 4.2ALLEE216 pKa = 3.88PTKK219 pKa = 10.43IYY221 pKa = 9.19THH223 pKa = 6.53LLEE226 pKa = 5.5PIYY229 pKa = 9.67EE230 pKa = 4.36TGVRR234 pKa = 11.84TAAHH238 pKa = 5.58ITGGGWTNIEE248 pKa = 3.67RR249 pKa = 11.84MGEE252 pKa = 4.06NTYY255 pKa = 10.42HH256 pKa = 6.41IHH258 pKa = 7.64DD259 pKa = 4.97PLSVPSIFEE268 pKa = 4.94LIQEE272 pKa = 4.62SGNIDD277 pKa = 3.65DD278 pKa = 4.55RR279 pKa = 11.84EE280 pKa = 4.15MYY282 pKa = 9.29RR283 pKa = 11.84TFNMGIGFVCAGEE296 pKa = 4.05PSVIDD301 pKa = 3.55SLVAEE306 pKa = 4.61TDD308 pKa = 3.64GEE310 pKa = 4.68VIGEE314 pKa = 4.12VDD316 pKa = 3.06VGTGVKK322 pKa = 9.26IDD324 pKa = 3.62GMVLL328 pKa = 3.2
MM1 pKa = 7.76TDD3 pKa = 3.61DD4 pKa = 4.65SSPSSDD10 pKa = 4.53EE11 pKa = 3.76EE12 pKa = 3.9LTYY15 pKa = 11.33AKK17 pKa = 10.54AGVDD21 pKa = 3.08IDD23 pKa = 4.05ASEE26 pKa = 4.3SATDD30 pKa = 3.84ALLQAVGEE38 pKa = 4.52TPGDD42 pKa = 3.58YY43 pKa = 10.89AGLIDD48 pKa = 5.7IGNQYY53 pKa = 10.86LAVTTDD59 pKa = 3.45GVGTKK64 pKa = 10.53LLIAEE69 pKa = 5.67AIAEE73 pKa = 4.14YY74 pKa = 9.31STIGIDD80 pKa = 3.56CVAMNVNDD88 pKa = 4.12LVAEE92 pKa = 4.65GVRR95 pKa = 11.84PIGFVDD101 pKa = 3.97YY102 pKa = 10.83LAVDD106 pKa = 3.94NPDD109 pKa = 3.4EE110 pKa = 5.85DD111 pKa = 3.52IASQIGVGLQSGADD125 pKa = 3.41RR126 pKa = 11.84ADD128 pKa = 2.91ISLIGGEE135 pKa = 4.16TAVMPEE141 pKa = 4.27VINGFDD147 pKa = 4.1LAGTCVGLAPKK158 pKa = 10.28SDD160 pKa = 3.64VLTQTAQPGDD170 pKa = 3.88IIAGFPSSGIHH181 pKa = 6.89SNGLTLARR189 pKa = 11.84KK190 pKa = 9.52AIQNQFSYY198 pKa = 10.26TDD200 pKa = 3.6QYY202 pKa = 11.6PEE204 pKa = 3.83SQYY207 pKa = 11.77NSIAEE212 pKa = 4.2ALLEE216 pKa = 3.88PTKK219 pKa = 10.43IYY221 pKa = 9.19THH223 pKa = 6.53LLEE226 pKa = 5.5PIYY229 pKa = 9.67EE230 pKa = 4.36TGVRR234 pKa = 11.84TAAHH238 pKa = 5.58ITGGGWTNIEE248 pKa = 3.67RR249 pKa = 11.84MGEE252 pKa = 4.06NTYY255 pKa = 10.42HH256 pKa = 6.41IHH258 pKa = 7.64DD259 pKa = 4.97PLSVPSIFEE268 pKa = 4.94LIQEE272 pKa = 4.62SGNIDD277 pKa = 3.65DD278 pKa = 4.55RR279 pKa = 11.84EE280 pKa = 4.15MYY282 pKa = 9.29RR283 pKa = 11.84TFNMGIGFVCAGEE296 pKa = 4.05PSVIDD301 pKa = 3.55SLVAEE306 pKa = 4.61TDD308 pKa = 3.64GEE310 pKa = 4.68VIGEE314 pKa = 4.12VDD316 pKa = 3.06VGTGVKK322 pKa = 9.26IDD324 pKa = 3.62GMVLL328 pKa = 3.2
Molecular weight: 34.55 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|U1Q140|U1Q140_9EURY 16S rRNA aminocarboxypropyltransferase OS=Haloquadratum sp. J07HQX50 OX=1238426 GN=J07HQX50_02621 PE=3 SV=1
MM1 pKa = 7.13QPLRR5 pKa = 11.84AYY7 pKa = 9.77SWRR10 pKa = 11.84EE11 pKa = 3.24TKK13 pKa = 9.99IYY15 pKa = 10.2YY16 pKa = 10.23DD17 pKa = 3.57RR18 pKa = 11.84FLSEE22 pKa = 4.63SRR24 pKa = 11.84PHH26 pKa = 6.88AQGQLNCISPGLTRR40 pKa = 11.84LQSIMCRR47 pKa = 11.84ALPKK51 pKa = 9.66FTGIFGWSDD60 pKa = 3.04SSTLSTATRR69 pKa = 11.84NSTALRR75 pKa = 11.84TKK77 pKa = 8.99TCEE80 pKa = 4.05SRR82 pKa = 11.84PTSQRR87 pKa = 11.84TACRR91 pKa = 11.84QLGRR95 pKa = 11.84WLLKK99 pKa = 10.14HH100 pKa = 5.93SRR102 pKa = 11.84RR103 pKa = 11.84SCASKK108 pKa = 10.36KK109 pKa = 10.76GKK111 pKa = 10.34LSANSRR117 pKa = 11.84FEE119 pKa = 4.41GLLSCMTTVTFNNDD133 pKa = 2.56HH134 pKa = 6.79CALSATDD141 pKa = 3.26QRR143 pKa = 11.84VRR145 pKa = 11.84AEE147 pKa = 3.69YY148 pKa = 10.58VRR150 pKa = 11.84RR151 pKa = 11.84HH152 pKa = 6.23DD153 pKa = 3.55SDD155 pKa = 3.77VVRR158 pKa = 11.84NRR160 pKa = 11.84LEE162 pKa = 3.64QSS164 pKa = 2.95
MM1 pKa = 7.13QPLRR5 pKa = 11.84AYY7 pKa = 9.77SWRR10 pKa = 11.84EE11 pKa = 3.24TKK13 pKa = 9.99IYY15 pKa = 10.2YY16 pKa = 10.23DD17 pKa = 3.57RR18 pKa = 11.84FLSEE22 pKa = 4.63SRR24 pKa = 11.84PHH26 pKa = 6.88AQGQLNCISPGLTRR40 pKa = 11.84LQSIMCRR47 pKa = 11.84ALPKK51 pKa = 9.66FTGIFGWSDD60 pKa = 3.04SSTLSTATRR69 pKa = 11.84NSTALRR75 pKa = 11.84TKK77 pKa = 8.99TCEE80 pKa = 4.05SRR82 pKa = 11.84PTSQRR87 pKa = 11.84TACRR91 pKa = 11.84QLGRR95 pKa = 11.84WLLKK99 pKa = 10.14HH100 pKa = 5.93SRR102 pKa = 11.84RR103 pKa = 11.84SCASKK108 pKa = 10.36KK109 pKa = 10.76GKK111 pKa = 10.34LSANSRR117 pKa = 11.84FEE119 pKa = 4.41GLLSCMTTVTFNNDD133 pKa = 2.56HH134 pKa = 6.79CALSATDD141 pKa = 3.26QRR143 pKa = 11.84VRR145 pKa = 11.84AEE147 pKa = 3.69YY148 pKa = 10.58VRR150 pKa = 11.84RR151 pKa = 11.84HH152 pKa = 6.23DD153 pKa = 3.55SDD155 pKa = 3.77VVRR158 pKa = 11.84NRR160 pKa = 11.84LEE162 pKa = 3.64QSS164 pKa = 2.95
Molecular weight: 18.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
653713 |
24 |
2413 |
228.2 |
24.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.141 ± 0.052 | 0.905 ± 0.017 |
6.763 ± 0.055 | 7.604 ± 0.061 |
3.359 ± 0.031 | 7.46 ± 0.059 |
2.237 ± 0.026 | 5.859 ± 0.041 |
2.411 ± 0.028 | 8.587 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.938 ± 0.022 | 3.189 ± 0.033 |
4.437 ± 0.032 | 3.729 ± 0.029 |
6.131 ± 0.05 | 7.192 ± 0.05 |
7.224 ± 0.05 | 7.872 ± 0.047 |
1.064 ± 0.018 | 2.742 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
