
Poecile atricapillus GI tract-associated gemycircularvirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus chickad1; Chickadee associated gemycircularvirus 1
Average proteome isoelectric point is 6.99
Get precalculated fractions of proteins
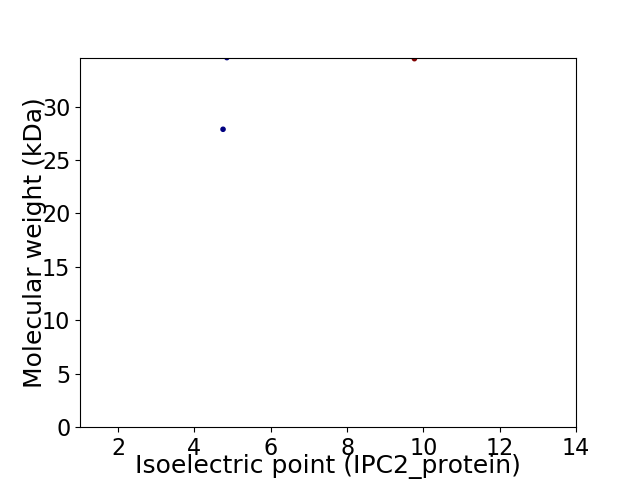
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K1P922|A0A0K1P922_9VIRU Putative capsid protein OS=Poecile atricapillus GI tract-associated gemycircularvirus OX=1699094 PE=4 SV=1
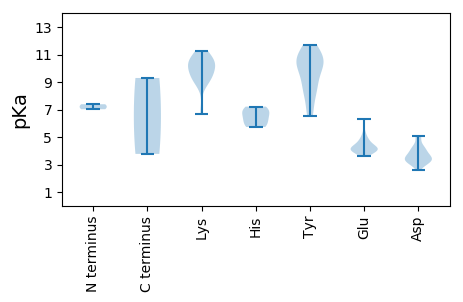
MM1 pKa = 7.39LGDD4 pKa = 3.52VGVLIVRR11 pKa = 11.84PVSCVLLYY19 pKa = 10.86NISFQQDD26 pKa = 3.36SEE28 pKa = 4.56DD29 pKa = 3.27TFMPSIHH36 pKa = 6.46FCAQHH41 pKa = 6.27FLLTYY46 pKa = 10.3AHH48 pKa = 6.86SEE50 pKa = 4.25GNDD53 pKa = 3.3VKK55 pKa = 11.21PEE57 pKa = 4.19LDD59 pKa = 3.07PFRR62 pKa = 11.84IVEE65 pKa = 4.06VLGALGAEE73 pKa = 4.39CIVARR78 pKa = 11.84EE79 pKa = 4.11YY80 pKa = 11.55YY81 pKa = 6.81PTTVGFHH88 pKa = 5.84LHH90 pKa = 5.73VFCSFEE96 pKa = 3.72RR97 pKa = 11.84RR98 pKa = 11.84FRR100 pKa = 11.84SRR102 pKa = 11.84KK103 pKa = 8.9IDD105 pKa = 3.35VFDD108 pKa = 3.49VDD110 pKa = 5.11GYY112 pKa = 11.15HH113 pKa = 7.21PNAEE117 pKa = 4.19PSRR120 pKa = 11.84KK121 pKa = 9.03NALGGYY127 pKa = 9.94DD128 pKa = 3.68YY129 pKa = 10.81AIKK132 pKa = 10.7DD133 pKa = 3.82GEE135 pKa = 4.57VVAGGLEE142 pKa = 4.15RR143 pKa = 11.84PSGVSGRR150 pKa = 11.84RR151 pKa = 11.84GPNRR155 pKa = 11.84SADD158 pKa = 3.07EE159 pKa = 3.96WAEE162 pKa = 3.85ITASEE167 pKa = 4.26TVEE170 pKa = 4.19EE171 pKa = 4.78FWRR174 pKa = 11.84LCEE177 pKa = 3.95EE178 pKa = 5.05LDD180 pKa = 4.39PKK182 pKa = 11.25SLVCNFPALLRR193 pKa = 11.84FSEE196 pKa = 4.01WRR198 pKa = 11.84FRR200 pKa = 11.84PVEE203 pKa = 3.99IPYY206 pKa = 8.14ATPDD210 pKa = 3.48GVFDD214 pKa = 4.17LAGYY218 pKa = 8.96PDD220 pKa = 3.83LADD223 pKa = 3.15WRR225 pKa = 11.84DD226 pKa = 3.3NVLFGGGSGEE236 pKa = 5.32SPFLWAEE243 pKa = 3.62QTKK246 pKa = 10.27IVV248 pKa = 3.77
MM1 pKa = 7.39LGDD4 pKa = 3.52VGVLIVRR11 pKa = 11.84PVSCVLLYY19 pKa = 10.86NISFQQDD26 pKa = 3.36SEE28 pKa = 4.56DD29 pKa = 3.27TFMPSIHH36 pKa = 6.46FCAQHH41 pKa = 6.27FLLTYY46 pKa = 10.3AHH48 pKa = 6.86SEE50 pKa = 4.25GNDD53 pKa = 3.3VKK55 pKa = 11.21PEE57 pKa = 4.19LDD59 pKa = 3.07PFRR62 pKa = 11.84IVEE65 pKa = 4.06VLGALGAEE73 pKa = 4.39CIVARR78 pKa = 11.84EE79 pKa = 4.11YY80 pKa = 11.55YY81 pKa = 6.81PTTVGFHH88 pKa = 5.84LHH90 pKa = 5.73VFCSFEE96 pKa = 3.72RR97 pKa = 11.84RR98 pKa = 11.84FRR100 pKa = 11.84SRR102 pKa = 11.84KK103 pKa = 8.9IDD105 pKa = 3.35VFDD108 pKa = 3.49VDD110 pKa = 5.11GYY112 pKa = 11.15HH113 pKa = 7.21PNAEE117 pKa = 4.19PSRR120 pKa = 11.84KK121 pKa = 9.03NALGGYY127 pKa = 9.94DD128 pKa = 3.68YY129 pKa = 10.81AIKK132 pKa = 10.7DD133 pKa = 3.82GEE135 pKa = 4.57VVAGGLEE142 pKa = 4.15RR143 pKa = 11.84PSGVSGRR150 pKa = 11.84RR151 pKa = 11.84GPNRR155 pKa = 11.84SADD158 pKa = 3.07EE159 pKa = 3.96WAEE162 pKa = 3.85ITASEE167 pKa = 4.26TVEE170 pKa = 4.19EE171 pKa = 4.78FWRR174 pKa = 11.84LCEE177 pKa = 3.95EE178 pKa = 5.05LDD180 pKa = 4.39PKK182 pKa = 11.25SLVCNFPALLRR193 pKa = 11.84FSEE196 pKa = 4.01WRR198 pKa = 11.84FRR200 pKa = 11.84PVEE203 pKa = 3.99IPYY206 pKa = 8.14ATPDD210 pKa = 3.48GVFDD214 pKa = 4.17LAGYY218 pKa = 8.96PDD220 pKa = 3.83LADD223 pKa = 3.15WRR225 pKa = 11.84DD226 pKa = 3.3NVLFGGGSGEE236 pKa = 5.32SPFLWAEE243 pKa = 3.62QTKK246 pKa = 10.27IVV248 pKa = 3.77
Molecular weight: 27.88 kDa
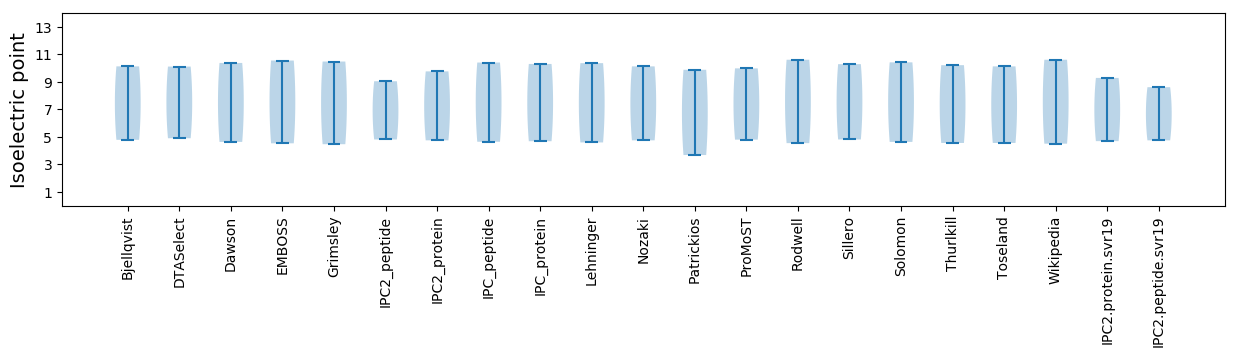
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K1P922|A0A0K1P922_9VIRU Putative capsid protein OS=Poecile atricapillus GI tract-associated gemycircularvirus OX=1699094 PE=4 SV=1
MM1 pKa = 7.02PRR3 pKa = 11.84KK4 pKa = 9.45YY5 pKa = 10.47LSRR8 pKa = 11.84YY9 pKa = 7.23RR10 pKa = 11.84PKK12 pKa = 10.72YY13 pKa = 8.16SGAYY17 pKa = 8.66RR18 pKa = 11.84GRR20 pKa = 11.84RR21 pKa = 11.84PVRR24 pKa = 11.84AKK26 pKa = 10.19ASPRR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84SYY34 pKa = 10.18GRR36 pKa = 11.84KK37 pKa = 8.97RR38 pKa = 11.84YY39 pKa = 9.45GGKK42 pKa = 6.68TTSKK46 pKa = 10.63KK47 pKa = 10.44RR48 pKa = 11.84ILNLTSIKK56 pKa = 10.55KK57 pKa = 9.7RR58 pKa = 11.84DD59 pKa = 3.49TMLAYY64 pKa = 9.58YY65 pKa = 10.48QNASTVNPQVGGYY78 pKa = 6.49TLQGTGGGNGGPATMLMWCATARR101 pKa = 11.84DD102 pKa = 3.86NTVAMGGSLGNRR114 pKa = 11.84FTPSTRR120 pKa = 11.84TATTCYY126 pKa = 9.76MRR128 pKa = 11.84GLSEE132 pKa = 4.69NIEE135 pKa = 4.12LSSNSGVPWQWRR147 pKa = 11.84RR148 pKa = 11.84ICFTFRR154 pKa = 11.84DD155 pKa = 4.17PASLSTVQSFYY166 pKa = 11.7NEE168 pKa = 4.15GTSGFQRR175 pKa = 11.84LLYY178 pKa = 10.26NLTSGSSPDD187 pKa = 3.52NVNIANVRR195 pKa = 11.84GLLFRR200 pKa = 11.84GEE202 pKa = 4.82LGQDD206 pKa = 2.61WASYY210 pKa = 8.31LTAPTDD216 pKa = 3.47NSRR219 pKa = 11.84VTIKK223 pKa = 9.75MDD225 pKa = 3.05KK226 pKa = 10.06TRR228 pKa = 11.84TISSGNQSGRR238 pKa = 11.84MVYY241 pKa = 10.57QKK243 pKa = 9.47MWMPMNKK250 pKa = 9.51NLVYY254 pKa = 10.83DD255 pKa = 4.2DD256 pKa = 5.05DD257 pKa = 4.91EE258 pKa = 6.32NGGSEE263 pKa = 3.98TARR266 pKa = 11.84FFSVGGKK273 pKa = 10.28AGMGDD278 pKa = 4.25YY279 pKa = 10.75YY280 pKa = 11.5VVDD283 pKa = 3.99IFQAGAGAVSSDD295 pKa = 3.08TLLFNPQATLYY306 pKa = 8.79WHH308 pKa = 6.98EE309 pKa = 4.28KK310 pKa = 9.3
MM1 pKa = 7.02PRR3 pKa = 11.84KK4 pKa = 9.45YY5 pKa = 10.47LSRR8 pKa = 11.84YY9 pKa = 7.23RR10 pKa = 11.84PKK12 pKa = 10.72YY13 pKa = 8.16SGAYY17 pKa = 8.66RR18 pKa = 11.84GRR20 pKa = 11.84RR21 pKa = 11.84PVRR24 pKa = 11.84AKK26 pKa = 10.19ASPRR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84SYY34 pKa = 10.18GRR36 pKa = 11.84KK37 pKa = 8.97RR38 pKa = 11.84YY39 pKa = 9.45GGKK42 pKa = 6.68TTSKK46 pKa = 10.63KK47 pKa = 10.44RR48 pKa = 11.84ILNLTSIKK56 pKa = 10.55KK57 pKa = 9.7RR58 pKa = 11.84DD59 pKa = 3.49TMLAYY64 pKa = 9.58YY65 pKa = 10.48QNASTVNPQVGGYY78 pKa = 6.49TLQGTGGGNGGPATMLMWCATARR101 pKa = 11.84DD102 pKa = 3.86NTVAMGGSLGNRR114 pKa = 11.84FTPSTRR120 pKa = 11.84TATTCYY126 pKa = 9.76MRR128 pKa = 11.84GLSEE132 pKa = 4.69NIEE135 pKa = 4.12LSSNSGVPWQWRR147 pKa = 11.84RR148 pKa = 11.84ICFTFRR154 pKa = 11.84DD155 pKa = 4.17PASLSTVQSFYY166 pKa = 11.7NEE168 pKa = 4.15GTSGFQRR175 pKa = 11.84LLYY178 pKa = 10.26NLTSGSSPDD187 pKa = 3.52NVNIANVRR195 pKa = 11.84GLLFRR200 pKa = 11.84GEE202 pKa = 4.82LGQDD206 pKa = 2.61WASYY210 pKa = 8.31LTAPTDD216 pKa = 3.47NSRR219 pKa = 11.84VTIKK223 pKa = 9.75MDD225 pKa = 3.05KK226 pKa = 10.06TRR228 pKa = 11.84TISSGNQSGRR238 pKa = 11.84MVYY241 pKa = 10.57QKK243 pKa = 9.47MWMPMNKK250 pKa = 9.51NLVYY254 pKa = 10.83DD255 pKa = 4.2DD256 pKa = 5.05DD257 pKa = 4.91EE258 pKa = 6.32NGGSEE263 pKa = 3.98TARR266 pKa = 11.84FFSVGGKK273 pKa = 10.28AGMGDD278 pKa = 4.25YY279 pKa = 10.75YY280 pKa = 11.5VVDD283 pKa = 3.99IFQAGAGAVSSDD295 pKa = 3.08TLLFNPQATLYY306 pKa = 8.79WHH308 pKa = 6.98EE309 pKa = 4.28KK310 pKa = 9.3
Molecular weight: 34.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
558 |
248 |
310 |
279.0 |
31.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.631 ± 0.15 | 1.613 ± 0.541 |
5.376 ± 0.991 | 5.018 ± 2.313 |
4.839 ± 1.352 | 9.857 ± 0.931 |
1.254 ± 0.781 | 3.226 ± 0.541 |
3.763 ± 0.901 | 7.527 ± 0.631 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.509 ± 1.142 | 4.659 ± 1.232 |
5.197 ± 0.841 | 2.688 ± 0.721 |
8.065 ± 0.811 | 8.065 ± 1.082 |
6.272 ± 2.043 | 6.631 ± 1.502 |
1.971 ± 0.03 | 4.839 ± 0.811 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
