
Coleophoma cylindrospora
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Leotiomycetes; Helotiales; Dermateaceae; Coleophoma
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
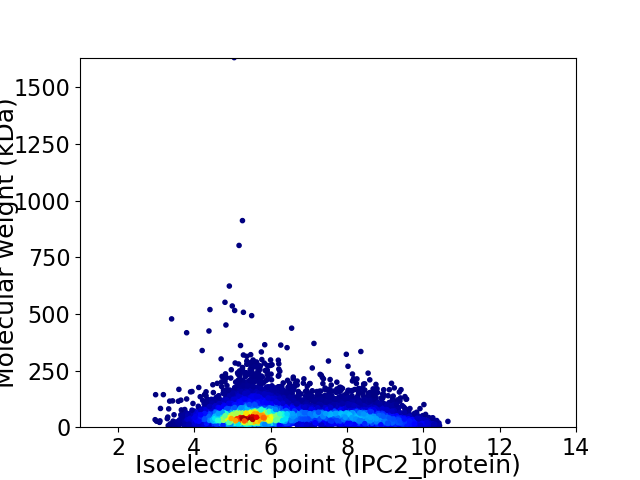
Virtual 2D-PAGE plot for 14176 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3D8RH41|A0A3D8RH41_9HELO Uncharacterized protein OS=Coleophoma cylindrospora OX=1849047 GN=BP6252_07173 PE=4 SV=1
MM1 pKa = 7.42ATPTTADD8 pKa = 2.86EE9 pKa = 4.06WHH11 pKa = 6.72TLDD14 pKa = 3.81SHH16 pKa = 6.94SPTRR20 pKa = 11.84NVATCPIDD28 pKa = 4.51DD29 pKa = 4.64LNEE32 pKa = 4.29NPTSAHH38 pKa = 5.96EE39 pKa = 4.21SQPAEE44 pKa = 4.31HH45 pKa = 6.52YY46 pKa = 10.23RR47 pKa = 11.84STMSNGNGVTHH58 pKa = 7.22RR59 pKa = 11.84FIDD62 pKa = 3.45QPSPHH67 pKa = 6.32EE68 pKa = 3.77QHH70 pKa = 6.86ASSEE74 pKa = 4.47CAYY77 pKa = 10.31MMSPYY82 pKa = 10.13TDD84 pKa = 5.33DD85 pKa = 3.72ISLEE89 pKa = 4.05EE90 pKa = 4.88DD91 pKa = 3.55LGQQSDD97 pKa = 4.19EE98 pKa = 4.31EE99 pKa = 4.14AVEE102 pKa = 4.08PGEE105 pKa = 4.65PLRR108 pKa = 11.84AWLTCMFISLADD120 pKa = 3.96PNGDD124 pKa = 3.57GIMISASQSSSSSLDD139 pKa = 3.36VEE141 pKa = 4.8LSDD144 pKa = 5.56SAGQHH149 pKa = 6.62PDD151 pKa = 3.12ATDD154 pKa = 3.61DD155 pKa = 4.36SAMSDD160 pKa = 3.62LNRR163 pKa = 11.84ATDD166 pKa = 3.56TTEE169 pKa = 4.02FPLMGIGSQFGASYY183 pKa = 10.28FIEE186 pKa = 4.43EE187 pKa = 4.17
MM1 pKa = 7.42ATPTTADD8 pKa = 2.86EE9 pKa = 4.06WHH11 pKa = 6.72TLDD14 pKa = 3.81SHH16 pKa = 6.94SPTRR20 pKa = 11.84NVATCPIDD28 pKa = 4.51DD29 pKa = 4.64LNEE32 pKa = 4.29NPTSAHH38 pKa = 5.96EE39 pKa = 4.21SQPAEE44 pKa = 4.31HH45 pKa = 6.52YY46 pKa = 10.23RR47 pKa = 11.84STMSNGNGVTHH58 pKa = 7.22RR59 pKa = 11.84FIDD62 pKa = 3.45QPSPHH67 pKa = 6.32EE68 pKa = 3.77QHH70 pKa = 6.86ASSEE74 pKa = 4.47CAYY77 pKa = 10.31MMSPYY82 pKa = 10.13TDD84 pKa = 5.33DD85 pKa = 3.72ISLEE89 pKa = 4.05EE90 pKa = 4.88DD91 pKa = 3.55LGQQSDD97 pKa = 4.19EE98 pKa = 4.31EE99 pKa = 4.14AVEE102 pKa = 4.08PGEE105 pKa = 4.65PLRR108 pKa = 11.84AWLTCMFISLADD120 pKa = 3.96PNGDD124 pKa = 3.57GIMISASQSSSSSLDD139 pKa = 3.36VEE141 pKa = 4.8LSDD144 pKa = 5.56SAGQHH149 pKa = 6.62PDD151 pKa = 3.12ATDD154 pKa = 3.61DD155 pKa = 4.36SAMSDD160 pKa = 3.62LNRR163 pKa = 11.84ATDD166 pKa = 3.56TTEE169 pKa = 4.02FPLMGIGSQFGASYY183 pKa = 10.28FIEE186 pKa = 4.43EE187 pKa = 4.17
Molecular weight: 20.27 kDa
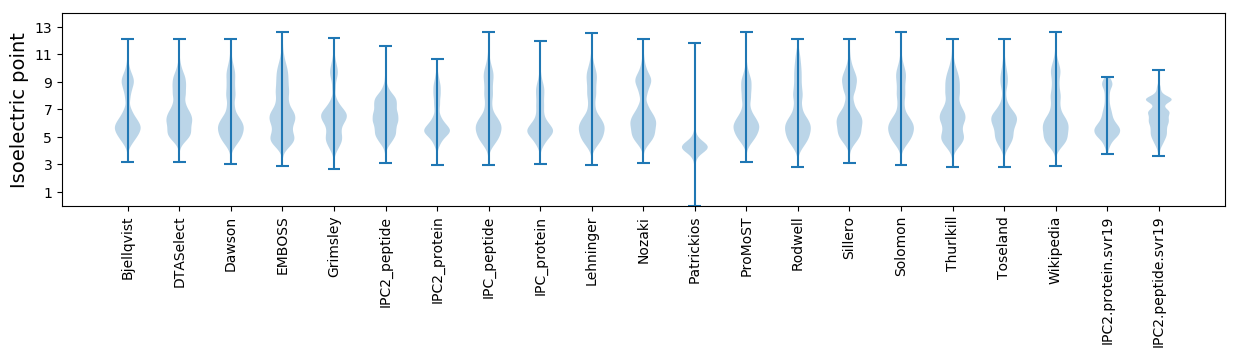
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3D8RN20|A0A3D8RN20_9HELO Uncharacterized protein OS=Coleophoma cylindrospora OX=1849047 GN=BP6252_06333 PE=4 SV=1
MM1 pKa = 7.51KK2 pKa = 9.98CAQAILQLVCILSFLYY18 pKa = 10.73NDD20 pKa = 3.17AHH22 pKa = 6.24AQVTVYY28 pKa = 9.55LTEE31 pKa = 4.31TTTATAPPVTVTIKK45 pKa = 9.81EE46 pKa = 4.4TGSCSWPTLMSNSITPSISTSNPTRR71 pKa = 11.84SSTPSLTSRR80 pKa = 11.84PFASSTIPPNSVQSTGTSLRR100 pKa = 11.84NSNSASPTSEE110 pKa = 3.91SLTNPSSLVHH120 pKa = 5.3STGTSLQTSNSASPVSEE137 pKa = 3.97SSTNPSSPVQSTGTSLRR154 pKa = 11.84ISNSASPASGSSTNPLSPVQPTGTSLRR181 pKa = 11.84TSNLASPASGSSSNPSSLVTRR202 pKa = 11.84TSLRR206 pKa = 11.84NSKK209 pKa = 10.1SASPVSGSSTNPSSLVQSTGTGLQTSNSASPASEE243 pKa = 3.94SLSNPSIPVQSTGTSLRR260 pKa = 11.84NSNSGSPTTGGSSNPSSLVTRR281 pKa = 11.84TSLQTSNSASLVSGSSTNPSSPVQSTRR308 pKa = 11.84TSLRR312 pKa = 11.84NSNSASPASGSSTNPSSPVQSPGTSQSTGTGQSPGTSQSPGTSQSPGTSQSPGTSLRR369 pKa = 11.84TSISPTPVSGSLTGTPLTSPGQTRR393 pKa = 11.84IISTPTNPSSSRR405 pKa = 11.84GPSSPATISTISGLSSRR422 pKa = 11.84SAQPSSIVSSQPPTRR437 pKa = 11.84PLTTSSSIKK446 pKa = 9.51TSSTKK451 pKa = 10.35TIIRR455 pKa = 11.84EE456 pKa = 4.16TKK458 pKa = 9.85RR459 pKa = 11.84PQSSASTVRR468 pKa = 11.84TSLLTTQPQLSSRR481 pKa = 11.84SSQISSSVSLTPSQLSSSGTRR502 pKa = 11.84TGSLPTQSQTGSSVSSILSQLSSSGTRR529 pKa = 11.84TGSIPTQSQTGSSVSSILSQLSSSGTRR556 pKa = 11.84TGSLPTQSQTGSSVSPILSQISSSGTRR583 pKa = 11.84TGSLPTQSQTGSSVSPISSQILRR606 pKa = 11.84SSVGTSSLPLQSQASSRR623 pKa = 11.84LSQTASSVPSTLSQISSDD641 pKa = 3.38TSTGSLPTQSQTGASASSILPQPSSSSIGTSSLPLQSQASSRR683 pKa = 11.84LAQTASSASLSTSGLQATSLPTGSQFSSGATSRR716 pKa = 11.84STAAAASSQSLISISSSAAQSNSLQATSSLTEE748 pKa = 4.4SPTSSPFTISPEE760 pKa = 4.17DD761 pKa = 3.4ASGASMSRR769 pKa = 11.84FSLSRR774 pKa = 11.84PGTSQTSAAPSGASTIPSSIGVSTLSSGLPSTTGSSIISMQGTSQSSSLNTQSPSRR830 pKa = 11.84ISTSEE835 pKa = 3.84TSILTPTAPGLSSTVTPALSQSSKK859 pKa = 10.76LGSTSSVSSAGGRR872 pKa = 11.84AGALGAVRR880 pKa = 11.84PGVVGATISRR890 pKa = 11.84ASTSPTTTLPTTTGSSGPSNSGSSLSSVPILSQPPSSIPMTATSNLLSPSTTLISTPSNSPLATPSTPLSSIPSIRR966 pKa = 11.84QSSTPGTGLSSVTIPGQSTTPGTLSPEE993 pKa = 4.19LSTTPGTPLSSTPIPGQSTTPGTPLSSALISGQSTTPGTPLSSALISGQSTTPGKK1048 pKa = 9.3PLSSTPIPGQSSTPGVTTPGLSTTPGTRR1076 pKa = 11.84PASTPIPGQSTIPGTPSPGQSTTHH1100 pKa = 5.73GTPNPGQPTTPGTPTPGQSTTPGNPTPGLSTTLGTASPGQSSTPGTQSPGMFVTPGTRR1158 pKa = 11.84PASTPVPGQSTTQGTPNPGQSTEE1181 pKa = 4.46PGTPSPGLSSSPSTPNSGQSTTPGTILASNPIPGQSTTQGTTLSSTPISGQSTTSGTPLFSTPILGQSSTPGAPSPGLSTTPGTRR1266 pKa = 11.84PASTPIPGQSTAPGTTNPGQSTTPGTPLSSTPIPGQSSAPGASSPGLSTTIPGQSSTPGAPSPGLSSTPGTLNPGQSTAPGTTNPGQSTTPGRR1359 pKa = 11.84PLSSTPIPGQSSTPGAPSPGLSTTPGTRR1387 pKa = 11.84LASTPTPGQSTTPGTPSSGQSTTLGTQLSSTPTPGQSTIPGTTLSSVPSASRR1439 pKa = 11.84STTSGAALTSTPGTAEE1455 pKa = 4.35LSTPSQSSLLPSGASPSLQASPSAISTSSVQSASASASTLSGASTPLSGSTGSSHH1510 pKa = 5.55STSVQPATSVSSFLTTFKK1528 pKa = 11.02SSTTSSPSATSTCVDD1543 pKa = 3.2RR1544 pKa = 11.84LVVNGGFEE1552 pKa = 4.21TGALSPWSQPVTLEE1566 pKa = 3.62VSAFFGPINSPTLCRR1581 pKa = 11.84TGQYY1585 pKa = 10.82CLGNGLYY1592 pKa = 10.42DD1593 pKa = 3.93GSNIMFKK1600 pKa = 10.57QSVQLCSNEE1609 pKa = 4.1SFRR1612 pKa = 11.84ASAWARR1618 pKa = 11.84ADD1620 pKa = 3.51TGDD1623 pKa = 3.71SVCYY1627 pKa = 10.08FEE1629 pKa = 6.37VCIQSPKK1636 pKa = 9.31HH1637 pKa = 5.8CSGPIIVDD1645 pKa = 3.8TTWTEE1650 pKa = 3.57ASFTFSGAPFSPKK1663 pKa = 10.2ADD1665 pKa = 3.37LEE1667 pKa = 4.69VYY1669 pKa = 8.81TGCIGDD1675 pKa = 4.44DD1676 pKa = 3.48STSFEE1681 pKa = 4.55IFIDD1685 pKa = 4.69DD1686 pKa = 4.27IAIQQVV1692 pKa = 2.92
MM1 pKa = 7.51KK2 pKa = 9.98CAQAILQLVCILSFLYY18 pKa = 10.73NDD20 pKa = 3.17AHH22 pKa = 6.24AQVTVYY28 pKa = 9.55LTEE31 pKa = 4.31TTTATAPPVTVTIKK45 pKa = 9.81EE46 pKa = 4.4TGSCSWPTLMSNSITPSISTSNPTRR71 pKa = 11.84SSTPSLTSRR80 pKa = 11.84PFASSTIPPNSVQSTGTSLRR100 pKa = 11.84NSNSASPTSEE110 pKa = 3.91SLTNPSSLVHH120 pKa = 5.3STGTSLQTSNSASPVSEE137 pKa = 3.97SSTNPSSPVQSTGTSLRR154 pKa = 11.84ISNSASPASGSSTNPLSPVQPTGTSLRR181 pKa = 11.84TSNLASPASGSSSNPSSLVTRR202 pKa = 11.84TSLRR206 pKa = 11.84NSKK209 pKa = 10.1SASPVSGSSTNPSSLVQSTGTGLQTSNSASPASEE243 pKa = 3.94SLSNPSIPVQSTGTSLRR260 pKa = 11.84NSNSGSPTTGGSSNPSSLVTRR281 pKa = 11.84TSLQTSNSASLVSGSSTNPSSPVQSTRR308 pKa = 11.84TSLRR312 pKa = 11.84NSNSASPASGSSTNPSSPVQSPGTSQSTGTGQSPGTSQSPGTSQSPGTSQSPGTSLRR369 pKa = 11.84TSISPTPVSGSLTGTPLTSPGQTRR393 pKa = 11.84IISTPTNPSSSRR405 pKa = 11.84GPSSPATISTISGLSSRR422 pKa = 11.84SAQPSSIVSSQPPTRR437 pKa = 11.84PLTTSSSIKK446 pKa = 9.51TSSTKK451 pKa = 10.35TIIRR455 pKa = 11.84EE456 pKa = 4.16TKK458 pKa = 9.85RR459 pKa = 11.84PQSSASTVRR468 pKa = 11.84TSLLTTQPQLSSRR481 pKa = 11.84SSQISSSVSLTPSQLSSSGTRR502 pKa = 11.84TGSLPTQSQTGSSVSSILSQLSSSGTRR529 pKa = 11.84TGSIPTQSQTGSSVSSILSQLSSSGTRR556 pKa = 11.84TGSLPTQSQTGSSVSPILSQISSSGTRR583 pKa = 11.84TGSLPTQSQTGSSVSPISSQILRR606 pKa = 11.84SSVGTSSLPLQSQASSRR623 pKa = 11.84LSQTASSVPSTLSQISSDD641 pKa = 3.38TSTGSLPTQSQTGASASSILPQPSSSSIGTSSLPLQSQASSRR683 pKa = 11.84LAQTASSASLSTSGLQATSLPTGSQFSSGATSRR716 pKa = 11.84STAAAASSQSLISISSSAAQSNSLQATSSLTEE748 pKa = 4.4SPTSSPFTISPEE760 pKa = 4.17DD761 pKa = 3.4ASGASMSRR769 pKa = 11.84FSLSRR774 pKa = 11.84PGTSQTSAAPSGASTIPSSIGVSTLSSGLPSTTGSSIISMQGTSQSSSLNTQSPSRR830 pKa = 11.84ISTSEE835 pKa = 3.84TSILTPTAPGLSSTVTPALSQSSKK859 pKa = 10.76LGSTSSVSSAGGRR872 pKa = 11.84AGALGAVRR880 pKa = 11.84PGVVGATISRR890 pKa = 11.84ASTSPTTTLPTTTGSSGPSNSGSSLSSVPILSQPPSSIPMTATSNLLSPSTTLISTPSNSPLATPSTPLSSIPSIRR966 pKa = 11.84QSSTPGTGLSSVTIPGQSTTPGTLSPEE993 pKa = 4.19LSTTPGTPLSSTPIPGQSTTPGTPLSSALISGQSTTPGTPLSSALISGQSTTPGKK1048 pKa = 9.3PLSSTPIPGQSSTPGVTTPGLSTTPGTRR1076 pKa = 11.84PASTPIPGQSTIPGTPSPGQSTTHH1100 pKa = 5.73GTPNPGQPTTPGTPTPGQSTTPGNPTPGLSTTLGTASPGQSSTPGTQSPGMFVTPGTRR1158 pKa = 11.84PASTPVPGQSTTQGTPNPGQSTEE1181 pKa = 4.46PGTPSPGLSSSPSTPNSGQSTTPGTILASNPIPGQSTTQGTTLSSTPISGQSTTSGTPLFSTPILGQSSTPGAPSPGLSTTPGTRR1266 pKa = 11.84PASTPIPGQSTAPGTTNPGQSTTPGTPLSSTPIPGQSSAPGASSPGLSTTIPGQSSTPGAPSPGLSSTPGTLNPGQSTAPGTTNPGQSTTPGRR1359 pKa = 11.84PLSSTPIPGQSSTPGAPSPGLSTTPGTRR1387 pKa = 11.84LASTPTPGQSTTPGTPSSGQSTTLGTQLSSTPTPGQSTIPGTTLSSVPSASRR1439 pKa = 11.84STTSGAALTSTPGTAEE1455 pKa = 4.35LSTPSQSSLLPSGASPSLQASPSAISTSSVQSASASASTLSGASTPLSGSTGSSHH1510 pKa = 5.55STSVQPATSVSSFLTTFKK1528 pKa = 11.02SSTTSSPSATSTCVDD1543 pKa = 3.2RR1544 pKa = 11.84LVVNGGFEE1552 pKa = 4.21TGALSPWSQPVTLEE1566 pKa = 3.62VSAFFGPINSPTLCRR1581 pKa = 11.84TGQYY1585 pKa = 10.82CLGNGLYY1592 pKa = 10.42DD1593 pKa = 3.93GSNIMFKK1600 pKa = 10.57QSVQLCSNEE1609 pKa = 4.1SFRR1612 pKa = 11.84ASAWARR1618 pKa = 11.84ADD1620 pKa = 3.51TGDD1623 pKa = 3.71SVCYY1627 pKa = 10.08FEE1629 pKa = 6.37VCIQSPKK1636 pKa = 9.31HH1637 pKa = 5.8CSGPIIVDD1645 pKa = 3.8TTWTEE1650 pKa = 3.57ASFTFSGAPFSPKK1663 pKa = 10.2ADD1665 pKa = 3.37LEE1667 pKa = 4.69VYY1669 pKa = 8.81TGCIGDD1675 pKa = 4.44DD1676 pKa = 3.48STSFEE1681 pKa = 4.55IFIDD1685 pKa = 4.69DD1686 pKa = 4.27IAIQQVV1692 pKa = 2.92
Molecular weight: 165.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7204061 |
49 |
14673 |
508.2 |
56.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.66 ± 0.019 | 1.262 ± 0.009 |
5.448 ± 0.015 | 6.022 ± 0.021 |
3.846 ± 0.011 | 6.945 ± 0.02 |
2.252 ± 0.009 | 5.326 ± 0.016 |
4.875 ± 0.018 | 8.961 ± 0.02 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.235 ± 0.008 | 3.818 ± 0.011 |
5.69 ± 0.019 | 3.927 ± 0.013 |
5.458 ± 0.019 | 8.459 ± 0.031 |
6.258 ± 0.023 | 6.151 ± 0.014 |
1.513 ± 0.008 | 2.896 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
