
Nitrosomonas sp. Nm58
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Nitrosomonadales; Nitrosomonadaceae; Nitrosomonas; unclassified Nitrosomonas
Average proteome isoelectric point is 6.73
Get precalculated fractions of proteins
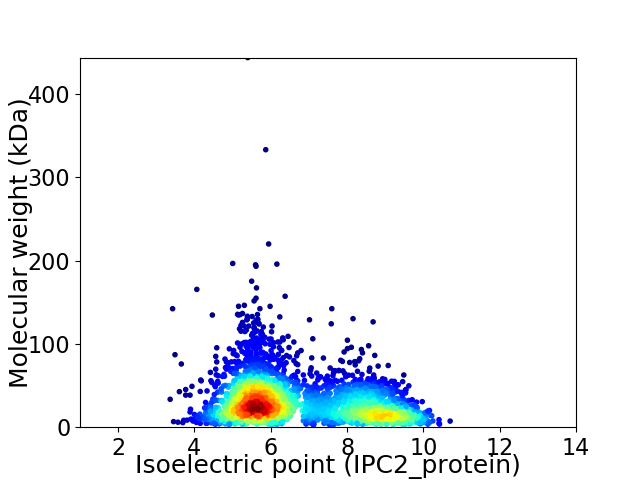
Virtual 2D-PAGE plot for 3100 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H3Q2D5|A0A1H3Q2D5_9PROT KfrB domain-containing protein OS=Nitrosomonas sp. Nm58 OX=200126 GN=SAMN05421754_105213 PE=4 SV=1
MM1 pKa = 7.65SNLLFNVTGSVDD13 pKa = 4.05DD14 pKa = 4.82DD15 pKa = 4.5LLIGSIGDD23 pKa = 4.71DD24 pKa = 3.06ILTGNLGNDD33 pKa = 3.57TLIGGVGNDD42 pKa = 3.13VAVYY46 pKa = 10.38FDD48 pKa = 4.21DD49 pKa = 3.53QANYY53 pKa = 9.95EE54 pKa = 4.05FSLNFSGQTTVRR66 pKa = 11.84DD67 pKa = 3.57LTTADD72 pKa = 3.63GDD74 pKa = 3.79EE75 pKa = 4.57GTDD78 pKa = 3.29VLLDD82 pKa = 3.37IEE84 pKa = 4.58IVRR87 pKa = 11.84FSDD90 pKa = 3.5GDD92 pKa = 3.73VKK94 pKa = 11.03LSYY97 pKa = 10.47QNSGEE102 pKa = 4.02FHH104 pKa = 6.85INTYY108 pKa = 6.57TTGDD112 pKa = 3.23QFAPEE117 pKa = 3.86ITALTNGGFVVTWTSEE133 pKa = 4.16GQDD136 pKa = 2.75GGAYY140 pKa = 9.79VIYY143 pKa = 9.5GQRR146 pKa = 11.84YY147 pKa = 7.54DD148 pKa = 3.8ANGVAQGNEE157 pKa = 3.94FLVNTTTTSSYY168 pKa = 10.53LPSVTTLTSGGFVVTWSSEE187 pKa = 3.9APGSKK192 pKa = 9.59FHH194 pKa = 7.8IYY196 pKa = 9.38GQRR199 pKa = 11.84FDD201 pKa = 5.15ANGITNGSEE210 pKa = 3.9FLVSTNTTHH219 pKa = 6.03SQSSSSITALTNGGFVVVWISDD241 pKa = 3.61AQDD244 pKa = 3.29GSDD247 pKa = 3.31YY248 pKa = 11.13GIYY251 pKa = 10.08GQRR254 pKa = 11.84FDD256 pKa = 5.34ANGAAQDD263 pKa = 4.12SEE265 pKa = 4.3FHH267 pKa = 6.78INTTTTTTYY276 pKa = 10.32SQSSPLITALTNGGFVVTWISEE298 pKa = 4.25GQDD301 pKa = 3.18SKK303 pKa = 11.66FDD305 pKa = 3.59IYY307 pKa = 10.26GQRR310 pKa = 11.84YY311 pKa = 7.59DD312 pKa = 3.86ANGIAQAGEE321 pKa = 3.88FRR323 pKa = 11.84VNTTTTTTSSLDD335 pKa = 3.42HH336 pKa = 6.92SIVALTNGGFVATWSSGSSSSDD358 pKa = 2.68GFGIYY363 pKa = 8.82GQRR366 pKa = 11.84FDD368 pKa = 4.25VNGVAQGNEE377 pKa = 3.94FLVNTTTTRR386 pKa = 11.84SEE388 pKa = 4.07SSPSITALTNGSFVITWSSVDD409 pKa = 3.36QDD411 pKa = 4.16GKK413 pKa = 10.84NDD415 pKa = 3.08IYY417 pKa = 11.0GQRR420 pKa = 11.84FDD422 pKa = 4.59ANGVAQGSEE431 pKa = 4.27FLVNTYY437 pKa = 8.77TFGNQSTSSTTALTNGGFVVTWVSTDD463 pKa = 3.39QEE465 pKa = 4.49SGDD468 pKa = 3.41VSDD471 pKa = 5.63RR472 pKa = 11.84FFSPGGIYY480 pKa = 9.23GQRR483 pKa = 11.84YY484 pKa = 9.16DD485 pKa = 4.12PEE487 pKa = 4.5GNKK490 pKa = 9.98IGDD493 pKa = 3.62LTLTGDD499 pKa = 3.61ANNNNLHH506 pKa = 5.98VADD509 pKa = 4.06TTIASIRR516 pKa = 11.84LLGMEE521 pKa = 4.71GNDD524 pKa = 3.06RR525 pKa = 11.84LQGSVGNDD533 pKa = 3.15ALDD536 pKa = 3.82GGIGADD542 pKa = 3.65TLIGGLGNDD551 pKa = 3.78TYY553 pKa = 11.31FVDD556 pKa = 3.68NVGDD560 pKa = 3.66IVTEE564 pKa = 4.09SLNAGTDD571 pKa = 3.77TVNSSIDD578 pKa = 3.37YY579 pKa = 7.41TLPTNLEE586 pKa = 4.17NLNMTGTLAINGTGNDD602 pKa = 3.97LDD604 pKa = 4.74NIIIANSADD613 pKa = 3.39NQLNGGAGNDD623 pKa = 3.85TLKK626 pKa = 10.85AGYY629 pKa = 9.92GRR631 pKa = 11.84DD632 pKa = 3.49VLTGGVGDD640 pKa = 5.36DD641 pKa = 3.34IFNFFAVGDD650 pKa = 4.05FVIHH654 pKa = 6.97DD655 pKa = 4.35FVNSSDD661 pKa = 4.18LLVFDD666 pKa = 5.04SKK668 pKa = 11.38ATGLYY673 pKa = 10.24DD674 pKa = 3.54INNLLSVISSIEE686 pKa = 3.96DD687 pKa = 3.26SSEE690 pKa = 3.52GAIVHH695 pKa = 6.84FIDD698 pKa = 4.84NIASITLIGLHH709 pKa = 6.63SDD711 pKa = 3.82DD712 pKa = 5.57LSTSMVGFAA721 pKa = 5.08
MM1 pKa = 7.65SNLLFNVTGSVDD13 pKa = 4.05DD14 pKa = 4.82DD15 pKa = 4.5LLIGSIGDD23 pKa = 4.71DD24 pKa = 3.06ILTGNLGNDD33 pKa = 3.57TLIGGVGNDD42 pKa = 3.13VAVYY46 pKa = 10.38FDD48 pKa = 4.21DD49 pKa = 3.53QANYY53 pKa = 9.95EE54 pKa = 4.05FSLNFSGQTTVRR66 pKa = 11.84DD67 pKa = 3.57LTTADD72 pKa = 3.63GDD74 pKa = 3.79EE75 pKa = 4.57GTDD78 pKa = 3.29VLLDD82 pKa = 3.37IEE84 pKa = 4.58IVRR87 pKa = 11.84FSDD90 pKa = 3.5GDD92 pKa = 3.73VKK94 pKa = 11.03LSYY97 pKa = 10.47QNSGEE102 pKa = 4.02FHH104 pKa = 6.85INTYY108 pKa = 6.57TTGDD112 pKa = 3.23QFAPEE117 pKa = 3.86ITALTNGGFVVTWTSEE133 pKa = 4.16GQDD136 pKa = 2.75GGAYY140 pKa = 9.79VIYY143 pKa = 9.5GQRR146 pKa = 11.84YY147 pKa = 7.54DD148 pKa = 3.8ANGVAQGNEE157 pKa = 3.94FLVNTTTTSSYY168 pKa = 10.53LPSVTTLTSGGFVVTWSSEE187 pKa = 3.9APGSKK192 pKa = 9.59FHH194 pKa = 7.8IYY196 pKa = 9.38GQRR199 pKa = 11.84FDD201 pKa = 5.15ANGITNGSEE210 pKa = 3.9FLVSTNTTHH219 pKa = 6.03SQSSSSITALTNGGFVVVWISDD241 pKa = 3.61AQDD244 pKa = 3.29GSDD247 pKa = 3.31YY248 pKa = 11.13GIYY251 pKa = 10.08GQRR254 pKa = 11.84FDD256 pKa = 5.34ANGAAQDD263 pKa = 4.12SEE265 pKa = 4.3FHH267 pKa = 6.78INTTTTTTYY276 pKa = 10.32SQSSPLITALTNGGFVVTWISEE298 pKa = 4.25GQDD301 pKa = 3.18SKK303 pKa = 11.66FDD305 pKa = 3.59IYY307 pKa = 10.26GQRR310 pKa = 11.84YY311 pKa = 7.59DD312 pKa = 3.86ANGIAQAGEE321 pKa = 3.88FRR323 pKa = 11.84VNTTTTTTSSLDD335 pKa = 3.42HH336 pKa = 6.92SIVALTNGGFVATWSSGSSSSDD358 pKa = 2.68GFGIYY363 pKa = 8.82GQRR366 pKa = 11.84FDD368 pKa = 4.25VNGVAQGNEE377 pKa = 3.94FLVNTTTTRR386 pKa = 11.84SEE388 pKa = 4.07SSPSITALTNGSFVITWSSVDD409 pKa = 3.36QDD411 pKa = 4.16GKK413 pKa = 10.84NDD415 pKa = 3.08IYY417 pKa = 11.0GQRR420 pKa = 11.84FDD422 pKa = 4.59ANGVAQGSEE431 pKa = 4.27FLVNTYY437 pKa = 8.77TFGNQSTSSTTALTNGGFVVTWVSTDD463 pKa = 3.39QEE465 pKa = 4.49SGDD468 pKa = 3.41VSDD471 pKa = 5.63RR472 pKa = 11.84FFSPGGIYY480 pKa = 9.23GQRR483 pKa = 11.84YY484 pKa = 9.16DD485 pKa = 4.12PEE487 pKa = 4.5GNKK490 pKa = 9.98IGDD493 pKa = 3.62LTLTGDD499 pKa = 3.61ANNNNLHH506 pKa = 5.98VADD509 pKa = 4.06TTIASIRR516 pKa = 11.84LLGMEE521 pKa = 4.71GNDD524 pKa = 3.06RR525 pKa = 11.84LQGSVGNDD533 pKa = 3.15ALDD536 pKa = 3.82GGIGADD542 pKa = 3.65TLIGGLGNDD551 pKa = 3.78TYY553 pKa = 11.31FVDD556 pKa = 3.68NVGDD560 pKa = 3.66IVTEE564 pKa = 4.09SLNAGTDD571 pKa = 3.77TVNSSIDD578 pKa = 3.37YY579 pKa = 7.41TLPTNLEE586 pKa = 4.17NLNMTGTLAINGTGNDD602 pKa = 3.97LDD604 pKa = 4.74NIIIANSADD613 pKa = 3.39NQLNGGAGNDD623 pKa = 3.85TLKK626 pKa = 10.85AGYY629 pKa = 9.92GRR631 pKa = 11.84DD632 pKa = 3.49VLTGGVGDD640 pKa = 5.36DD641 pKa = 3.34IFNFFAVGDD650 pKa = 4.05FVIHH654 pKa = 6.97DD655 pKa = 4.35FVNSSDD661 pKa = 4.18LLVFDD666 pKa = 5.04SKK668 pKa = 11.38ATGLYY673 pKa = 10.24DD674 pKa = 3.54INNLLSVISSIEE686 pKa = 3.96DD687 pKa = 3.26SSEE690 pKa = 3.52GAIVHH695 pKa = 6.84FIDD698 pKa = 4.84NIASITLIGLHH709 pKa = 6.63SDD711 pKa = 3.82DD712 pKa = 5.57LSTSMVGFAA721 pKa = 5.08
Molecular weight: 75.96 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H3LBM7|A0A1H3LBM7_9PROT Fatty acid desaturase OS=Nitrosomonas sp. Nm58 OX=200126 GN=SAMN05421754_101523 PE=4 SV=1
MM1 pKa = 7.76SADD4 pKa = 4.79QPLAWYY10 pKa = 6.82PVPVTASINIRR21 pKa = 11.84WWLLHH26 pKa = 6.69RR27 pKa = 11.84DD28 pKa = 3.35SLTRR32 pKa = 11.84LIQQRR37 pKa = 11.84CSKK40 pKa = 10.57FSVDD44 pKa = 4.94PLFQSLAPACTDD56 pKa = 3.23EE57 pKa = 4.53LVMMNLRR64 pKa = 11.84RR65 pKa = 11.84GEE67 pKa = 3.92LALVRR72 pKa = 11.84EE73 pKa = 4.58VYY75 pKa = 9.93LYY77 pKa = 11.07CGDD80 pKa = 3.63IPVVFAHH87 pKa = 5.82SVVARR92 pKa = 11.84KK93 pKa = 9.05HH94 pKa = 5.76LQSAWRR100 pKa = 11.84GLSRR104 pKa = 11.84LGNRR108 pKa = 11.84SLGTMLFTNPMIKK121 pKa = 9.75RR122 pKa = 11.84AEE124 pKa = 3.98FGYY127 pKa = 10.15KK128 pKa = 9.15QLNIHH133 pKa = 6.39HH134 pKa = 7.2PLFKK138 pKa = 10.27RR139 pKa = 11.84ACQRR143 pKa = 11.84LQTPPTSLWARR154 pKa = 11.84RR155 pKa = 11.84SLFTLQGQPILVTEE169 pKa = 4.27VFLPAILRR177 pKa = 11.84LKK179 pKa = 10.2II180 pKa = 3.73
MM1 pKa = 7.76SADD4 pKa = 4.79QPLAWYY10 pKa = 6.82PVPVTASINIRR21 pKa = 11.84WWLLHH26 pKa = 6.69RR27 pKa = 11.84DD28 pKa = 3.35SLTRR32 pKa = 11.84LIQQRR37 pKa = 11.84CSKK40 pKa = 10.57FSVDD44 pKa = 4.94PLFQSLAPACTDD56 pKa = 3.23EE57 pKa = 4.53LVMMNLRR64 pKa = 11.84RR65 pKa = 11.84GEE67 pKa = 3.92LALVRR72 pKa = 11.84EE73 pKa = 4.58VYY75 pKa = 9.93LYY77 pKa = 11.07CGDD80 pKa = 3.63IPVVFAHH87 pKa = 5.82SVVARR92 pKa = 11.84KK93 pKa = 9.05HH94 pKa = 5.76LQSAWRR100 pKa = 11.84GLSRR104 pKa = 11.84LGNRR108 pKa = 11.84SLGTMLFTNPMIKK121 pKa = 9.75RR122 pKa = 11.84AEE124 pKa = 3.98FGYY127 pKa = 10.15KK128 pKa = 9.15QLNIHH133 pKa = 6.39HH134 pKa = 7.2PLFKK138 pKa = 10.27RR139 pKa = 11.84ACQRR143 pKa = 11.84LQTPPTSLWARR154 pKa = 11.84RR155 pKa = 11.84SLFTLQGQPILVTEE169 pKa = 4.27VFLPAILRR177 pKa = 11.84LKK179 pKa = 10.2II180 pKa = 3.73
Molecular weight: 20.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
926182 |
25 |
3980 |
298.8 |
33.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.099 ± 0.048 | 1.011 ± 0.016 |
5.135 ± 0.032 | 6.024 ± 0.042 |
4.051 ± 0.034 | 6.956 ± 0.044 |
2.525 ± 0.022 | 6.823 ± 0.042 |
4.699 ± 0.041 | 10.757 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.484 ± 0.024 | 3.841 ± 0.03 |
4.432 ± 0.03 | 4.222 ± 0.031 |
5.807 ± 0.037 | 6.126 ± 0.037 |
5.258 ± 0.033 | 6.493 ± 0.033 |
1.351 ± 0.02 | 2.905 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
