
Point-Douro narna-like virus
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 9.29
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Z2RSZ2|A0A1Z2RSZ2_9VIRU RdRp OS=Point-Douro narna-like virus OX=2010279 PE=4 SV=1
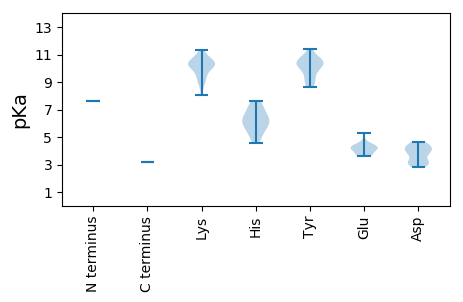
MM1 pKa = 7.63PLVARR6 pKa = 11.84PTSMRR11 pKa = 11.84VATRR15 pKa = 11.84SEE17 pKa = 3.73VSRR20 pKa = 11.84RR21 pKa = 11.84VNLAAEE27 pKa = 4.13TWKK30 pKa = 10.84AICAAVATRR39 pKa = 11.84KK40 pKa = 8.85WPKK43 pKa = 10.22FNLDD47 pKa = 3.18FQTDD51 pKa = 3.16RR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84FCTFQALARR63 pKa = 11.84HH64 pKa = 6.16LLLSALARR72 pKa = 11.84GPVEE76 pKa = 3.65TIKK79 pKa = 10.61RR80 pKa = 11.84VKK82 pKa = 10.03SWTAQIRR89 pKa = 11.84EE90 pKa = 4.23DD91 pKa = 3.68VFLHH95 pKa = 6.17GRR97 pKa = 11.84CPPSAFRR104 pKa = 11.84GLVGALGWTDD114 pKa = 3.78ANSHH118 pKa = 6.05LQLSYY123 pKa = 10.46IGRR126 pKa = 11.84ALPYY130 pKa = 10.29GDD132 pKa = 4.04ATICEE137 pKa = 4.48HH138 pKa = 6.53ALKK141 pKa = 10.66SHH143 pKa = 6.7ADD145 pKa = 3.47VLGSLGVTPPSLLGAARR162 pKa = 11.84GWAKK166 pKa = 10.22EE167 pKa = 3.61WAIKK171 pKa = 10.41NRR173 pKa = 11.84LSLDD177 pKa = 4.5LIRR180 pKa = 11.84EE181 pKa = 4.07PPIQEE186 pKa = 4.21SSCLEE191 pKa = 3.64YY192 pKa = 10.69SRR194 pKa = 11.84RR195 pKa = 11.84EE196 pKa = 3.74GGYY199 pKa = 9.14ARR201 pKa = 11.84AIKK204 pKa = 10.05EE205 pKa = 3.78HH206 pKa = 5.53TARR209 pKa = 11.84CLTLPPSSVRR219 pKa = 11.84HH220 pKa = 4.84SMKK223 pKa = 10.33EE224 pKa = 3.68GTRR227 pKa = 11.84ASNPGRR233 pKa = 11.84VLKK236 pKa = 10.77DD237 pKa = 3.05LAFADD242 pKa = 4.43LSNTARR248 pKa = 11.84VVAVPEE254 pKa = 4.4RR255 pKa = 11.84GWKK258 pKa = 9.77ARR260 pKa = 11.84IVTAHH265 pKa = 6.52AAARR269 pKa = 11.84VAFLHH274 pKa = 5.63SLRR277 pKa = 11.84HH278 pKa = 5.23GLFAALRR285 pKa = 11.84NDD287 pKa = 3.01RR288 pKa = 11.84RR289 pKa = 11.84TKK291 pKa = 10.35LVAEE295 pKa = 4.53GLHH298 pKa = 5.97RR299 pKa = 11.84EE300 pKa = 4.0AVEE303 pKa = 3.92ALFATGVDD311 pKa = 3.79PRR313 pKa = 11.84CTIVSADD320 pKa = 3.98LSAASDD326 pKa = 3.8TLHH329 pKa = 7.27GDD331 pKa = 4.04LLQAIVEE338 pKa = 4.33GLGDD342 pKa = 4.41ALGCTSKK349 pKa = 10.96FMDD352 pKa = 4.24EE353 pKa = 4.11FTNAAVGSYY362 pKa = 8.87TLSYY366 pKa = 10.3PDD368 pKa = 4.05GSTVTTQRR376 pKa = 11.84GALMGLPTTWPLLCLTHH393 pKa = 7.07LFWCDD398 pKa = 3.08QSLVQLHH405 pKa = 6.09GRR407 pKa = 11.84GGCRR411 pKa = 11.84EE412 pKa = 3.95KK413 pKa = 10.13EE414 pKa = 4.42TPGSSRR420 pKa = 11.84EE421 pKa = 4.1AEE423 pKa = 4.62VICGDD428 pKa = 4.27DD429 pKa = 4.46LAAAWRR435 pKa = 11.84PDD437 pKa = 2.92RR438 pKa = 11.84VAAYY442 pKa = 9.93EE443 pKa = 4.34EE444 pKa = 4.33IAVLCGAVFSPGKK457 pKa = 9.36HH458 pKa = 5.8LKK460 pKa = 10.23SKK462 pKa = 10.57VYY464 pKa = 10.63GIFTEE469 pKa = 5.28DD470 pKa = 3.17IYY472 pKa = 11.43SVQVSCVPTRR482 pKa = 11.84VTRR485 pKa = 11.84EE486 pKa = 3.69VSKK489 pKa = 9.14TWTGPVDD496 pKa = 4.03KK497 pKa = 11.33VPMNLVNEE505 pKa = 4.77LGQLLRR511 pKa = 11.84WRR513 pKa = 11.84AQRR516 pKa = 11.84YY517 pKa = 8.61KK518 pKa = 10.66VGKK521 pKa = 8.04TGAFQFPPRR530 pKa = 11.84NVTLSYY536 pKa = 10.72VSKK539 pKa = 9.72EE540 pKa = 4.18TGLKK544 pKa = 8.59MNPVSVLFRR553 pKa = 11.84EE554 pKa = 4.35WSTAIPLRR562 pKa = 11.84WAVRR566 pKa = 11.84APKK569 pKa = 9.47HH570 pKa = 5.31APGMRR575 pKa = 11.84GDD577 pKa = 4.2LPPWVTVPLAAHH589 pKa = 6.87AVASAHH595 pKa = 6.33PGRR598 pKa = 11.84WSKK601 pKa = 10.25VCRR604 pKa = 11.84VVKK607 pKa = 10.5LCYY610 pKa = 9.67PGMAKK615 pKa = 10.21FFASHH620 pKa = 7.38GIPPYY625 pKa = 10.31LPRR628 pKa = 11.84VLGGGGLPTPLGDD641 pKa = 2.98RR642 pKa = 11.84VKK644 pKa = 10.26IGRR647 pKa = 11.84VASRR651 pKa = 11.84RR652 pKa = 11.84IRR654 pKa = 11.84KK655 pKa = 8.95ALGGALYY662 pKa = 10.49RR663 pKa = 11.84STDD666 pKa = 3.12PTTFGSIWTTALSPAYY682 pKa = 10.15ALALADD688 pKa = 4.35AQDD691 pKa = 4.5LDD693 pKa = 4.66AMHH696 pKa = 7.6PDD698 pKa = 3.26AFKK701 pKa = 11.01VRR703 pKa = 11.84QTRR706 pKa = 11.84SLRR709 pKa = 11.84PPFKK713 pKa = 11.06DD714 pKa = 2.81NGNYY718 pKa = 10.35ADD720 pKa = 4.1FVEE723 pKa = 4.46MRR725 pKa = 11.84AGKK728 pKa = 10.38GSLWALRR735 pKa = 11.84QGAQLVGRR743 pKa = 11.84SFTPSPARR751 pKa = 11.84VSNALRR757 pKa = 11.84TKK759 pKa = 9.92VRR761 pKa = 11.84KK762 pKa = 9.37ALAKK766 pKa = 10.0RR767 pKa = 11.84GYY769 pKa = 9.25LRR771 pKa = 11.84SSAPVSKK778 pKa = 10.46LIEE781 pKa = 4.25RR782 pKa = 11.84ASVKK786 pKa = 10.32PSDD789 pKa = 3.7HH790 pKa = 4.59MWIRR794 pKa = 11.84EE795 pKa = 3.95RR796 pKa = 11.84PVLSFGRR803 pKa = 11.84FGGRR807 pKa = 11.84MSGKK811 pKa = 9.82KK812 pKa = 9.45EE813 pKa = 4.0EE814 pKa = 4.28PVSFQWKK821 pKa = 9.09KK822 pKa = 10.44RR823 pKa = 11.84EE824 pKa = 4.14SVKK827 pKa = 10.32TGSS830 pKa = 3.2
MM1 pKa = 7.63PLVARR6 pKa = 11.84PTSMRR11 pKa = 11.84VATRR15 pKa = 11.84SEE17 pKa = 3.73VSRR20 pKa = 11.84RR21 pKa = 11.84VNLAAEE27 pKa = 4.13TWKK30 pKa = 10.84AICAAVATRR39 pKa = 11.84KK40 pKa = 8.85WPKK43 pKa = 10.22FNLDD47 pKa = 3.18FQTDD51 pKa = 3.16RR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84FCTFQALARR63 pKa = 11.84HH64 pKa = 6.16LLLSALARR72 pKa = 11.84GPVEE76 pKa = 3.65TIKK79 pKa = 10.61RR80 pKa = 11.84VKK82 pKa = 10.03SWTAQIRR89 pKa = 11.84EE90 pKa = 4.23DD91 pKa = 3.68VFLHH95 pKa = 6.17GRR97 pKa = 11.84CPPSAFRR104 pKa = 11.84GLVGALGWTDD114 pKa = 3.78ANSHH118 pKa = 6.05LQLSYY123 pKa = 10.46IGRR126 pKa = 11.84ALPYY130 pKa = 10.29GDD132 pKa = 4.04ATICEE137 pKa = 4.48HH138 pKa = 6.53ALKK141 pKa = 10.66SHH143 pKa = 6.7ADD145 pKa = 3.47VLGSLGVTPPSLLGAARR162 pKa = 11.84GWAKK166 pKa = 10.22EE167 pKa = 3.61WAIKK171 pKa = 10.41NRR173 pKa = 11.84LSLDD177 pKa = 4.5LIRR180 pKa = 11.84EE181 pKa = 4.07PPIQEE186 pKa = 4.21SSCLEE191 pKa = 3.64YY192 pKa = 10.69SRR194 pKa = 11.84RR195 pKa = 11.84EE196 pKa = 3.74GGYY199 pKa = 9.14ARR201 pKa = 11.84AIKK204 pKa = 10.05EE205 pKa = 3.78HH206 pKa = 5.53TARR209 pKa = 11.84CLTLPPSSVRR219 pKa = 11.84HH220 pKa = 4.84SMKK223 pKa = 10.33EE224 pKa = 3.68GTRR227 pKa = 11.84ASNPGRR233 pKa = 11.84VLKK236 pKa = 10.77DD237 pKa = 3.05LAFADD242 pKa = 4.43LSNTARR248 pKa = 11.84VVAVPEE254 pKa = 4.4RR255 pKa = 11.84GWKK258 pKa = 9.77ARR260 pKa = 11.84IVTAHH265 pKa = 6.52AAARR269 pKa = 11.84VAFLHH274 pKa = 5.63SLRR277 pKa = 11.84HH278 pKa = 5.23GLFAALRR285 pKa = 11.84NDD287 pKa = 3.01RR288 pKa = 11.84RR289 pKa = 11.84TKK291 pKa = 10.35LVAEE295 pKa = 4.53GLHH298 pKa = 5.97RR299 pKa = 11.84EE300 pKa = 4.0AVEE303 pKa = 3.92ALFATGVDD311 pKa = 3.79PRR313 pKa = 11.84CTIVSADD320 pKa = 3.98LSAASDD326 pKa = 3.8TLHH329 pKa = 7.27GDD331 pKa = 4.04LLQAIVEE338 pKa = 4.33GLGDD342 pKa = 4.41ALGCTSKK349 pKa = 10.96FMDD352 pKa = 4.24EE353 pKa = 4.11FTNAAVGSYY362 pKa = 8.87TLSYY366 pKa = 10.3PDD368 pKa = 4.05GSTVTTQRR376 pKa = 11.84GALMGLPTTWPLLCLTHH393 pKa = 7.07LFWCDD398 pKa = 3.08QSLVQLHH405 pKa = 6.09GRR407 pKa = 11.84GGCRR411 pKa = 11.84EE412 pKa = 3.95KK413 pKa = 10.13EE414 pKa = 4.42TPGSSRR420 pKa = 11.84EE421 pKa = 4.1AEE423 pKa = 4.62VICGDD428 pKa = 4.27DD429 pKa = 4.46LAAAWRR435 pKa = 11.84PDD437 pKa = 2.92RR438 pKa = 11.84VAAYY442 pKa = 9.93EE443 pKa = 4.34EE444 pKa = 4.33IAVLCGAVFSPGKK457 pKa = 9.36HH458 pKa = 5.8LKK460 pKa = 10.23SKK462 pKa = 10.57VYY464 pKa = 10.63GIFTEE469 pKa = 5.28DD470 pKa = 3.17IYY472 pKa = 11.43SVQVSCVPTRR482 pKa = 11.84VTRR485 pKa = 11.84EE486 pKa = 3.69VSKK489 pKa = 9.14TWTGPVDD496 pKa = 4.03KK497 pKa = 11.33VPMNLVNEE505 pKa = 4.77LGQLLRR511 pKa = 11.84WRR513 pKa = 11.84AQRR516 pKa = 11.84YY517 pKa = 8.61KK518 pKa = 10.66VGKK521 pKa = 8.04TGAFQFPPRR530 pKa = 11.84NVTLSYY536 pKa = 10.72VSKK539 pKa = 9.72EE540 pKa = 4.18TGLKK544 pKa = 8.59MNPVSVLFRR553 pKa = 11.84EE554 pKa = 4.35WSTAIPLRR562 pKa = 11.84WAVRR566 pKa = 11.84APKK569 pKa = 9.47HH570 pKa = 5.31APGMRR575 pKa = 11.84GDD577 pKa = 4.2LPPWVTVPLAAHH589 pKa = 6.87AVASAHH595 pKa = 6.33PGRR598 pKa = 11.84WSKK601 pKa = 10.25VCRR604 pKa = 11.84VVKK607 pKa = 10.5LCYY610 pKa = 9.67PGMAKK615 pKa = 10.21FFASHH620 pKa = 7.38GIPPYY625 pKa = 10.31LPRR628 pKa = 11.84VLGGGGLPTPLGDD641 pKa = 2.98RR642 pKa = 11.84VKK644 pKa = 10.26IGRR647 pKa = 11.84VASRR651 pKa = 11.84RR652 pKa = 11.84IRR654 pKa = 11.84KK655 pKa = 8.95ALGGALYY662 pKa = 10.49RR663 pKa = 11.84STDD666 pKa = 3.12PTTFGSIWTTALSPAYY682 pKa = 10.15ALALADD688 pKa = 4.35AQDD691 pKa = 4.5LDD693 pKa = 4.66AMHH696 pKa = 7.6PDD698 pKa = 3.26AFKK701 pKa = 11.01VRR703 pKa = 11.84QTRR706 pKa = 11.84SLRR709 pKa = 11.84PPFKK713 pKa = 11.06DD714 pKa = 2.81NGNYY718 pKa = 10.35ADD720 pKa = 4.1FVEE723 pKa = 4.46MRR725 pKa = 11.84AGKK728 pKa = 10.38GSLWALRR735 pKa = 11.84QGAQLVGRR743 pKa = 11.84SFTPSPARR751 pKa = 11.84VSNALRR757 pKa = 11.84TKK759 pKa = 9.92VRR761 pKa = 11.84KK762 pKa = 9.37ALAKK766 pKa = 10.0RR767 pKa = 11.84GYY769 pKa = 9.25LRR771 pKa = 11.84SSAPVSKK778 pKa = 10.46LIEE781 pKa = 4.25RR782 pKa = 11.84ASVKK786 pKa = 10.32PSDD789 pKa = 3.7HH790 pKa = 4.59MWIRR794 pKa = 11.84EE795 pKa = 3.95RR796 pKa = 11.84PVLSFGRR803 pKa = 11.84FGGRR807 pKa = 11.84MSGKK811 pKa = 9.82KK812 pKa = 9.45EE813 pKa = 4.0EE814 pKa = 4.28PVSFQWKK821 pKa = 9.09KK822 pKa = 10.44RR823 pKa = 11.84EE824 pKa = 4.14SVKK827 pKa = 10.32TGSS830 pKa = 3.2
Molecular weight: 91.14 kDa
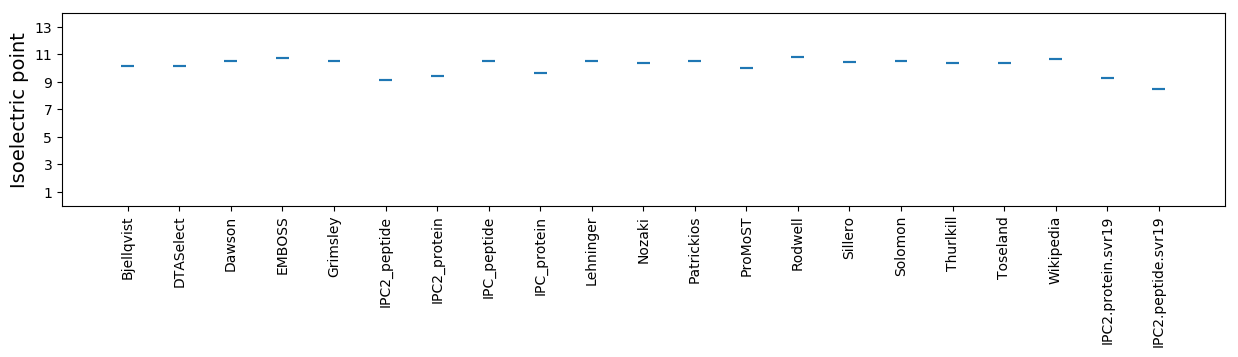
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Z2RSZ2|A0A1Z2RSZ2_9VIRU RdRp OS=Point-Douro narna-like virus OX=2010279 PE=4 SV=1
MM1 pKa = 7.63PLVARR6 pKa = 11.84PTSMRR11 pKa = 11.84VATRR15 pKa = 11.84SEE17 pKa = 3.73VSRR20 pKa = 11.84RR21 pKa = 11.84VNLAAEE27 pKa = 4.13TWKK30 pKa = 10.84AICAAVATRR39 pKa = 11.84KK40 pKa = 8.85WPKK43 pKa = 10.22FNLDD47 pKa = 3.18FQTDD51 pKa = 3.16RR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84FCTFQALARR63 pKa = 11.84HH64 pKa = 6.16LLLSALARR72 pKa = 11.84GPVEE76 pKa = 3.65TIKK79 pKa = 10.61RR80 pKa = 11.84VKK82 pKa = 10.03SWTAQIRR89 pKa = 11.84EE90 pKa = 4.23DD91 pKa = 3.68VFLHH95 pKa = 6.17GRR97 pKa = 11.84CPPSAFRR104 pKa = 11.84GLVGALGWTDD114 pKa = 3.78ANSHH118 pKa = 6.05LQLSYY123 pKa = 10.46IGRR126 pKa = 11.84ALPYY130 pKa = 10.29GDD132 pKa = 4.04ATICEE137 pKa = 4.48HH138 pKa = 6.53ALKK141 pKa = 10.66SHH143 pKa = 6.7ADD145 pKa = 3.47VLGSLGVTPPSLLGAARR162 pKa = 11.84GWAKK166 pKa = 10.22EE167 pKa = 3.61WAIKK171 pKa = 10.41NRR173 pKa = 11.84LSLDD177 pKa = 4.5LIRR180 pKa = 11.84EE181 pKa = 4.07PPIQEE186 pKa = 4.21SSCLEE191 pKa = 3.64YY192 pKa = 10.69SRR194 pKa = 11.84RR195 pKa = 11.84EE196 pKa = 3.74GGYY199 pKa = 9.14ARR201 pKa = 11.84AIKK204 pKa = 10.05EE205 pKa = 3.78HH206 pKa = 5.53TARR209 pKa = 11.84CLTLPPSSVRR219 pKa = 11.84HH220 pKa = 4.84SMKK223 pKa = 10.33EE224 pKa = 3.68GTRR227 pKa = 11.84ASNPGRR233 pKa = 11.84VLKK236 pKa = 10.77DD237 pKa = 3.05LAFADD242 pKa = 4.43LSNTARR248 pKa = 11.84VVAVPEE254 pKa = 4.4RR255 pKa = 11.84GWKK258 pKa = 9.77ARR260 pKa = 11.84IVTAHH265 pKa = 6.52AAARR269 pKa = 11.84VAFLHH274 pKa = 5.63SLRR277 pKa = 11.84HH278 pKa = 5.23GLFAALRR285 pKa = 11.84NDD287 pKa = 3.01RR288 pKa = 11.84RR289 pKa = 11.84TKK291 pKa = 10.35LVAEE295 pKa = 4.53GLHH298 pKa = 5.97RR299 pKa = 11.84EE300 pKa = 4.0AVEE303 pKa = 3.92ALFATGVDD311 pKa = 3.79PRR313 pKa = 11.84CTIVSADD320 pKa = 3.98LSAASDD326 pKa = 3.8TLHH329 pKa = 7.27GDD331 pKa = 4.04LLQAIVEE338 pKa = 4.33GLGDD342 pKa = 4.41ALGCTSKK349 pKa = 10.96FMDD352 pKa = 4.24EE353 pKa = 4.11FTNAAVGSYY362 pKa = 8.87TLSYY366 pKa = 10.3PDD368 pKa = 4.05GSTVTTQRR376 pKa = 11.84GALMGLPTTWPLLCLTHH393 pKa = 7.07LFWCDD398 pKa = 3.08QSLVQLHH405 pKa = 6.09GRR407 pKa = 11.84GGCRR411 pKa = 11.84EE412 pKa = 3.95KK413 pKa = 10.13EE414 pKa = 4.42TPGSSRR420 pKa = 11.84EE421 pKa = 4.1AEE423 pKa = 4.62VICGDD428 pKa = 4.27DD429 pKa = 4.46LAAAWRR435 pKa = 11.84PDD437 pKa = 2.92RR438 pKa = 11.84VAAYY442 pKa = 9.93EE443 pKa = 4.34EE444 pKa = 4.33IAVLCGAVFSPGKK457 pKa = 9.36HH458 pKa = 5.8LKK460 pKa = 10.23SKK462 pKa = 10.57VYY464 pKa = 10.63GIFTEE469 pKa = 5.28DD470 pKa = 3.17IYY472 pKa = 11.43SVQVSCVPTRR482 pKa = 11.84VTRR485 pKa = 11.84EE486 pKa = 3.69VSKK489 pKa = 9.14TWTGPVDD496 pKa = 4.03KK497 pKa = 11.33VPMNLVNEE505 pKa = 4.77LGQLLRR511 pKa = 11.84WRR513 pKa = 11.84AQRR516 pKa = 11.84YY517 pKa = 8.61KK518 pKa = 10.66VGKK521 pKa = 8.04TGAFQFPPRR530 pKa = 11.84NVTLSYY536 pKa = 10.72VSKK539 pKa = 9.72EE540 pKa = 4.18TGLKK544 pKa = 8.59MNPVSVLFRR553 pKa = 11.84EE554 pKa = 4.35WSTAIPLRR562 pKa = 11.84WAVRR566 pKa = 11.84APKK569 pKa = 9.47HH570 pKa = 5.31APGMRR575 pKa = 11.84GDD577 pKa = 4.2LPPWVTVPLAAHH589 pKa = 6.87AVASAHH595 pKa = 6.33PGRR598 pKa = 11.84WSKK601 pKa = 10.25VCRR604 pKa = 11.84VVKK607 pKa = 10.5LCYY610 pKa = 9.67PGMAKK615 pKa = 10.21FFASHH620 pKa = 7.38GIPPYY625 pKa = 10.31LPRR628 pKa = 11.84VLGGGGLPTPLGDD641 pKa = 2.98RR642 pKa = 11.84VKK644 pKa = 10.26IGRR647 pKa = 11.84VASRR651 pKa = 11.84RR652 pKa = 11.84IRR654 pKa = 11.84KK655 pKa = 8.95ALGGALYY662 pKa = 10.49RR663 pKa = 11.84STDD666 pKa = 3.12PTTFGSIWTTALSPAYY682 pKa = 10.15ALALADD688 pKa = 4.35AQDD691 pKa = 4.5LDD693 pKa = 4.66AMHH696 pKa = 7.6PDD698 pKa = 3.26AFKK701 pKa = 11.01VRR703 pKa = 11.84QTRR706 pKa = 11.84SLRR709 pKa = 11.84PPFKK713 pKa = 11.06DD714 pKa = 2.81NGNYY718 pKa = 10.35ADD720 pKa = 4.1FVEE723 pKa = 4.46MRR725 pKa = 11.84AGKK728 pKa = 10.38GSLWALRR735 pKa = 11.84QGAQLVGRR743 pKa = 11.84SFTPSPARR751 pKa = 11.84VSNALRR757 pKa = 11.84TKK759 pKa = 9.92VRR761 pKa = 11.84KK762 pKa = 9.37ALAKK766 pKa = 10.0RR767 pKa = 11.84GYY769 pKa = 9.25LRR771 pKa = 11.84SSAPVSKK778 pKa = 10.46LIEE781 pKa = 4.25RR782 pKa = 11.84ASVKK786 pKa = 10.32PSDD789 pKa = 3.7HH790 pKa = 4.59MWIRR794 pKa = 11.84EE795 pKa = 3.95RR796 pKa = 11.84PVLSFGRR803 pKa = 11.84FGGRR807 pKa = 11.84MSGKK811 pKa = 9.82KK812 pKa = 9.45EE813 pKa = 4.0EE814 pKa = 4.28PVSFQWKK821 pKa = 9.09KK822 pKa = 10.44RR823 pKa = 11.84EE824 pKa = 4.14SVKK827 pKa = 10.32TGSS830 pKa = 3.2
MM1 pKa = 7.63PLVARR6 pKa = 11.84PTSMRR11 pKa = 11.84VATRR15 pKa = 11.84SEE17 pKa = 3.73VSRR20 pKa = 11.84RR21 pKa = 11.84VNLAAEE27 pKa = 4.13TWKK30 pKa = 10.84AICAAVATRR39 pKa = 11.84KK40 pKa = 8.85WPKK43 pKa = 10.22FNLDD47 pKa = 3.18FQTDD51 pKa = 3.16RR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84FCTFQALARR63 pKa = 11.84HH64 pKa = 6.16LLLSALARR72 pKa = 11.84GPVEE76 pKa = 3.65TIKK79 pKa = 10.61RR80 pKa = 11.84VKK82 pKa = 10.03SWTAQIRR89 pKa = 11.84EE90 pKa = 4.23DD91 pKa = 3.68VFLHH95 pKa = 6.17GRR97 pKa = 11.84CPPSAFRR104 pKa = 11.84GLVGALGWTDD114 pKa = 3.78ANSHH118 pKa = 6.05LQLSYY123 pKa = 10.46IGRR126 pKa = 11.84ALPYY130 pKa = 10.29GDD132 pKa = 4.04ATICEE137 pKa = 4.48HH138 pKa = 6.53ALKK141 pKa = 10.66SHH143 pKa = 6.7ADD145 pKa = 3.47VLGSLGVTPPSLLGAARR162 pKa = 11.84GWAKK166 pKa = 10.22EE167 pKa = 3.61WAIKK171 pKa = 10.41NRR173 pKa = 11.84LSLDD177 pKa = 4.5LIRR180 pKa = 11.84EE181 pKa = 4.07PPIQEE186 pKa = 4.21SSCLEE191 pKa = 3.64YY192 pKa = 10.69SRR194 pKa = 11.84RR195 pKa = 11.84EE196 pKa = 3.74GGYY199 pKa = 9.14ARR201 pKa = 11.84AIKK204 pKa = 10.05EE205 pKa = 3.78HH206 pKa = 5.53TARR209 pKa = 11.84CLTLPPSSVRR219 pKa = 11.84HH220 pKa = 4.84SMKK223 pKa = 10.33EE224 pKa = 3.68GTRR227 pKa = 11.84ASNPGRR233 pKa = 11.84VLKK236 pKa = 10.77DD237 pKa = 3.05LAFADD242 pKa = 4.43LSNTARR248 pKa = 11.84VVAVPEE254 pKa = 4.4RR255 pKa = 11.84GWKK258 pKa = 9.77ARR260 pKa = 11.84IVTAHH265 pKa = 6.52AAARR269 pKa = 11.84VAFLHH274 pKa = 5.63SLRR277 pKa = 11.84HH278 pKa = 5.23GLFAALRR285 pKa = 11.84NDD287 pKa = 3.01RR288 pKa = 11.84RR289 pKa = 11.84TKK291 pKa = 10.35LVAEE295 pKa = 4.53GLHH298 pKa = 5.97RR299 pKa = 11.84EE300 pKa = 4.0AVEE303 pKa = 3.92ALFATGVDD311 pKa = 3.79PRR313 pKa = 11.84CTIVSADD320 pKa = 3.98LSAASDD326 pKa = 3.8TLHH329 pKa = 7.27GDD331 pKa = 4.04LLQAIVEE338 pKa = 4.33GLGDD342 pKa = 4.41ALGCTSKK349 pKa = 10.96FMDD352 pKa = 4.24EE353 pKa = 4.11FTNAAVGSYY362 pKa = 8.87TLSYY366 pKa = 10.3PDD368 pKa = 4.05GSTVTTQRR376 pKa = 11.84GALMGLPTTWPLLCLTHH393 pKa = 7.07LFWCDD398 pKa = 3.08QSLVQLHH405 pKa = 6.09GRR407 pKa = 11.84GGCRR411 pKa = 11.84EE412 pKa = 3.95KK413 pKa = 10.13EE414 pKa = 4.42TPGSSRR420 pKa = 11.84EE421 pKa = 4.1AEE423 pKa = 4.62VICGDD428 pKa = 4.27DD429 pKa = 4.46LAAAWRR435 pKa = 11.84PDD437 pKa = 2.92RR438 pKa = 11.84VAAYY442 pKa = 9.93EE443 pKa = 4.34EE444 pKa = 4.33IAVLCGAVFSPGKK457 pKa = 9.36HH458 pKa = 5.8LKK460 pKa = 10.23SKK462 pKa = 10.57VYY464 pKa = 10.63GIFTEE469 pKa = 5.28DD470 pKa = 3.17IYY472 pKa = 11.43SVQVSCVPTRR482 pKa = 11.84VTRR485 pKa = 11.84EE486 pKa = 3.69VSKK489 pKa = 9.14TWTGPVDD496 pKa = 4.03KK497 pKa = 11.33VPMNLVNEE505 pKa = 4.77LGQLLRR511 pKa = 11.84WRR513 pKa = 11.84AQRR516 pKa = 11.84YY517 pKa = 8.61KK518 pKa = 10.66VGKK521 pKa = 8.04TGAFQFPPRR530 pKa = 11.84NVTLSYY536 pKa = 10.72VSKK539 pKa = 9.72EE540 pKa = 4.18TGLKK544 pKa = 8.59MNPVSVLFRR553 pKa = 11.84EE554 pKa = 4.35WSTAIPLRR562 pKa = 11.84WAVRR566 pKa = 11.84APKK569 pKa = 9.47HH570 pKa = 5.31APGMRR575 pKa = 11.84GDD577 pKa = 4.2LPPWVTVPLAAHH589 pKa = 6.87AVASAHH595 pKa = 6.33PGRR598 pKa = 11.84WSKK601 pKa = 10.25VCRR604 pKa = 11.84VVKK607 pKa = 10.5LCYY610 pKa = 9.67PGMAKK615 pKa = 10.21FFASHH620 pKa = 7.38GIPPYY625 pKa = 10.31LPRR628 pKa = 11.84VLGGGGLPTPLGDD641 pKa = 2.98RR642 pKa = 11.84VKK644 pKa = 10.26IGRR647 pKa = 11.84VASRR651 pKa = 11.84RR652 pKa = 11.84IRR654 pKa = 11.84KK655 pKa = 8.95ALGGALYY662 pKa = 10.49RR663 pKa = 11.84STDD666 pKa = 3.12PTTFGSIWTTALSPAYY682 pKa = 10.15ALALADD688 pKa = 4.35AQDD691 pKa = 4.5LDD693 pKa = 4.66AMHH696 pKa = 7.6PDD698 pKa = 3.26AFKK701 pKa = 11.01VRR703 pKa = 11.84QTRR706 pKa = 11.84SLRR709 pKa = 11.84PPFKK713 pKa = 11.06DD714 pKa = 2.81NGNYY718 pKa = 10.35ADD720 pKa = 4.1FVEE723 pKa = 4.46MRR725 pKa = 11.84AGKK728 pKa = 10.38GSLWALRR735 pKa = 11.84QGAQLVGRR743 pKa = 11.84SFTPSPARR751 pKa = 11.84VSNALRR757 pKa = 11.84TKK759 pKa = 9.92VRR761 pKa = 11.84KK762 pKa = 9.37ALAKK766 pKa = 10.0RR767 pKa = 11.84GYY769 pKa = 9.25LRR771 pKa = 11.84SSAPVSKK778 pKa = 10.46LIEE781 pKa = 4.25RR782 pKa = 11.84ASVKK786 pKa = 10.32PSDD789 pKa = 3.7HH790 pKa = 4.59MWIRR794 pKa = 11.84EE795 pKa = 3.95RR796 pKa = 11.84PVLSFGRR803 pKa = 11.84FGGRR807 pKa = 11.84MSGKK811 pKa = 9.82KK812 pKa = 9.45EE813 pKa = 4.0EE814 pKa = 4.28PVSFQWKK821 pKa = 9.09KK822 pKa = 10.44RR823 pKa = 11.84EE824 pKa = 4.14SVKK827 pKa = 10.32TGSS830 pKa = 3.2
Molecular weight: 91.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
830 |
830 |
830 |
830.0 |
91.14 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.325 ± 0.0 | 1.928 ± 0.0 |
3.976 ± 0.0 | 4.217 ± 0.0 |
3.373 ± 0.0 | 7.831 ± 0.0 |
2.53 ± 0.0 | 2.771 ± 0.0 |
5.181 ± 0.0 | 10.0 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.566 ± 0.0 | 1.807 ± 0.0 |
6.265 ± 0.0 | 2.169 ± 0.0 |
9.398 ± 0.0 | 7.47 ± 0.0 |
6.024 ± 0.0 | 7.711 ± 0.0 |
2.41 ± 0.0 | 2.048 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
